Reaction: Autocatalytic phosphorylation of FGFR1 mutants with enhanced kinase activity
- in pathway: Signaling by activated point mutants of FGFR1
The three kinase domain mutants of FGFR1 that have been identified in glioblastoma are predicted or have been shown to result in enhanced kinase activity. The N546K (Rand, 2005) residue lies in a stretch of 9 amino acids that are conserved between all four FGFRs. Mutation of the paralogous residue in FGFR3 (N540K) has been shown to result in weak ligand-independent contstitutive activation in the autosomal disorder hypochodroplasia (Raffioni, 1998). In FGFR2 mutation of the paralogous residue to lysine has been identified in endometrial cancer and been shown to result in enhanced kinase activity (Dutt, 2008; Pollock, 2008); germline mutations at this site in FGFR2 are also associated with the development of Crouzon and Pfeiffer syndromes (Kan, 2002). The FGFR1 N546K mutations has accelerated rates of autophosphorylation and supports transformation when transfected into Rat-1 cells (Lew, 2009).
The FGFR1 K656E (TCGA, 2008) mutation is paralogous to activating mutations in FGFR3 kinase domain associated with the development of thanatophoric dysplasias (Tavormina, 1999; Bellus, 2000; Hart, 2000), and has itself been shown to activating when expressed in neural crest cells (Petiot, 2002).
The FGFR1 R576W (Rand, 2005) mutation increases the hydrophobicity of the receptor, and is postulated to enhance protein-protein interactions and thereby increase the likelihood of autophosphorylation of adjacent tyrosine residues, although this has not been explicitly demonstrated.
The FGFR1 K656E (TCGA, 2008) mutation is paralogous to activating mutations in FGFR3 kinase domain associated with the development of thanatophoric dysplasias (Tavormina, 1999; Bellus, 2000; Hart, 2000), and has itself been shown to activating when expressed in neural crest cells (Petiot, 2002).
The FGFR1 R576W (Rand, 2005) mutation increases the hydrophobicity of the receptor, and is postulated to enhance protein-protein interactions and thereby increase the likelihood of autophosphorylation of adjacent tyrosine residues, although this has not been explicitly demonstrated.
Reaction - small molecule participants:
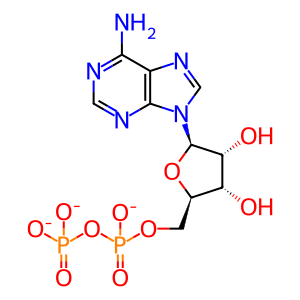
ADP [cytosol]
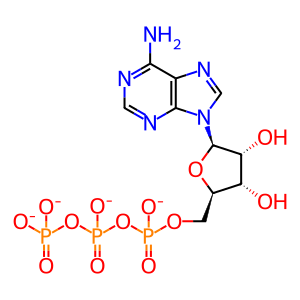
ATP [cytosol]
Reactome.org reaction link: R-HSA-2023460
======
Reaction input - small molecules:
ATP(4-)
Reaction output - small molecules:
ADP(3-)
Reactome.org link: R-HSA-2023460