Reaction: Activated TGFBR1 cannot phosphorylate SMAD2 and SMAD3 Phosphorylation Motif Mutants
- in pathway: SMAD2/3 Phosphorylation Motif Mutants in Cancer
SMAD2 and SMAD3 phosphorylation motif mutants cannot be phosphorylated and activated by the TGF-beta receptor complex (Fleming et al. 2013), in contrast to the wild-type SMAD2 and SMAD3 (Souchelnytskyi et al. 2001).
SMAD2 phosphorylation motif mutant SMAD2 S464*, which lacks the phosphorylation motif, has been annotated as a characterized member of the mutant set (Fleming et al. 2013), while SMAD2 S464L, SMAD S467P and SMAD3 S425C are candidate mutant set members. In addition, the following SMAD2 and SMAD3 truncation mutants that lack various portions of the MH2 domain, including phosphorylation sites, are annotated as candidate loss-of-function mutants:
SMAD2 S296*
SMAD2 S306*
SMAD2 S308*
SMAD2 L315*
SMAD2 R321*
SMAD2 W422*
SMAD2 R427*
SMAD2 Q429*
SMAD2 E439*
SMAD2 W448*
SMAD2 Q455*
SMAD2 C463*
SMAD3 E246*
SMAD3 Y298*
SMAD3 Q316*
SMAD3 Q322*
SMAD3 W326*
SMAD3 Q358*
SMAD3 R368*
SMAD3 E383*
SMAD3 Y384*
As SMAD2 and SMAD3 phosphorylation motif missense mutants (Fleming et al. 2013) have all the MH2 domain residues involved in ZFYVE9 (SARA)-mediated recruitment of SMAD2/3 to activated TGF-beta receptor complex 1 (TGFBR1) (Wu et al. 2000) intact, it is assumed that SMAD2/3 phosphorylation mutants bind phosphorylated TGFBR1. This has not been experimentally examined. It is likely that at least some of the nonsense SMAD2 and SMAD3 mutants, which lack parts of the MH2 domain, are unable to bind to ZFYVE9.
SMAD2 phosphorylation motif mutant SMAD2 S464*, which lacks the phosphorylation motif, has been annotated as a characterized member of the mutant set (Fleming et al. 2013), while SMAD2 S464L, SMAD S467P and SMAD3 S425C are candidate mutant set members. In addition, the following SMAD2 and SMAD3 truncation mutants that lack various portions of the MH2 domain, including phosphorylation sites, are annotated as candidate loss-of-function mutants:
SMAD2 S296*
SMAD2 S306*
SMAD2 S308*
SMAD2 L315*
SMAD2 R321*
SMAD2 W422*
SMAD2 R427*
SMAD2 Q429*
SMAD2 E439*
SMAD2 W448*
SMAD2 Q455*
SMAD2 C463*
SMAD3 E246*
SMAD3 Y298*
SMAD3 Q316*
SMAD3 Q322*
SMAD3 W326*
SMAD3 Q358*
SMAD3 R368*
SMAD3 E383*
SMAD3 Y384*
As SMAD2 and SMAD3 phosphorylation motif missense mutants (Fleming et al. 2013) have all the MH2 domain residues involved in ZFYVE9 (SARA)-mediated recruitment of SMAD2/3 to activated TGF-beta receptor complex 1 (TGFBR1) (Wu et al. 2000) intact, it is assumed that SMAD2/3 phosphorylation mutants bind phosphorylated TGFBR1. This has not been experimentally examined. It is likely that at least some of the nonsense SMAD2 and SMAD3 mutants, which lack parts of the MH2 domain, are unable to bind to ZFYVE9.
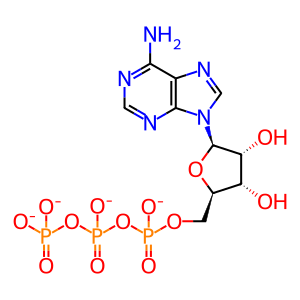
Reaction - small molecule participants:
ATP [cytosol]
Reactome.org reaction link: R-HSA-3304394
======
Reaction input - small molecules:
ATP(4-)
Reaction output - small molecules:
Reactome.org link: R-HSA-3304394