Reaction: ATP hydrolysis by HSP70
Heat shock protein 70 (HSP70) proteins bind and release client polypeptides in a cycle of cochaperone-mediated conformational changes that is coupled to ATP binding and hydrolysis (Mayer MP 2013). The overall domain structure of all HSP70 chaperone proteins is evolutionary conserved: the N-terminal nucleotide-binding domain (NBD) with ATPase activity is joined by a flexible linker to the C-terminal polypeptide substrate-binding domain (SBD). Most of our mechanistic understanding of HSP70 structure and function has come from analyses of Escherichia coli HSP70 family member, DnaK (Pellecchia M et al. 2000; Schuermann, JP et al. 2008; Bertelsen EB et al. 2009; Kityk R et al. 2012; Qi R et al. 2013). Chaperone function of bacterial DnaK involves an allosteric control mechanism between its two functional domains NBD and SBD. ATP binding and hydrolysis modulates the affinity of bacterial HSP70 protein for polypeptides, and polypeptide binding stimulates ATP hydrolysis (Mayer MP et al. 2000; Kityk R et al. 2012; Qi R et al. 2013). Also in the ATP-bound form, the lid domain remains open, which facilitates transient interactions with substrates. Following ATP hydrolysis, a conformational change releases the SBD, resulting in closure of the lid and a ~10-fold increase in the affinity for substrate (Wittung-Stafshede P et al. 2003; Slepenkov SV & Witt SN 2002). The conformation change associated with ATP hydrolysis is communicated through a key proline switch and involves the conserved, hydrophobic linker that connects the NBD to the SBD (Vogel M et al. 2006; Swain JF et al. 2007). ATP hydrolysis is essential for HSP70 chaperones, but the intrinsic ATPase rate is very low (Chang L et al. 2008). This ATPase activity of HSP70 is stimulated by protein substrates in synergism with J domain cochaperones (HSP40s) (Karzai AW & McMacken R 1996; Russell R et al. 1999; Laufen T et al. 1999; Landry SJ 2003; Wittung-Stafshede P et al. 2003).
The HSP70 family of chaperone proteins is one of the most conserved protein families in evolution (Takayama S et al. 1999; Boorstein WR et al. 1994; Brocchieri L et al. 2008). The sequence alignment of eukaryotic and bacterial HSP70 proteins revealed that the human HSP70 SBD is highly homologous to the DnaK SBD (51% sequence identity in the full-length protein and 47% identity in the SBD) (Zhang P et al. 2014). Moreover, the crystal structure of the substrate-bound human HSP70-SBD resembled the overall fold of the corresponding domain in the substrate-bound DnaK structures, confirming a similar overall architecture of the orthologous bacterial and human HSP70 proteins (Zhang P et al. 2014). Structures of nucleotide-binding domains of four human HSP70 isoforms: HSPA1L, HSPA2, HSPA6 and HSPA5 also support the view that the NBDs of human HSP70 function by conserved mechanisms (Wisniewska M et al. 2014). Structural analysis of a functionally intact bovine Hsp70 family member Hsc70 together with analysis of mutants in the interdomain linker and interface support the allosteric mechanism of the mammalian HSP70 chaperones (Wilbanks SM & McKay DB 1998; Jiang J et al. 2005).
Reaction - small molecule participants:
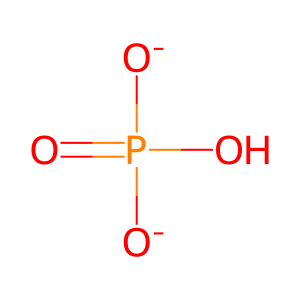
Pi [cytosol]
======
Reaction input - small molecules:
Reaction output - small molecules:
hydrogenphosphate