Reaction: Exosome Complex hydrolyzes mRNA by 3' to 5' exoribonuclease digestion
- in pathway: mRNA decay by 3' to 5' exoribonuclease
The exosome complex hydrolyzes capped, deadenylated mRNA from 3' to 5' and yields ribonucleotides having 5'-monophosphates. In yeast the Ski2-Ski3-Ski8 complex assists degradation by the exosome complex, however little is known about the function of the homologous Ski complex in mammals. Although many exosomal components contain exonuclease signatures, only two components have been shown to degrade RNA. Rrp6/PMSCL-100 has been shown to be involved in the 3’-5’ decay of nuclear mRNAs in yeast. Rrp6 may also function in the absence of the core exosomal components. The Rrp44/dDis3 component of the core exosome has been shown to possess both 3’-5’ exonuclease activity along with endonuclease activity via its PIN domain.
Reaction - small molecule participants:
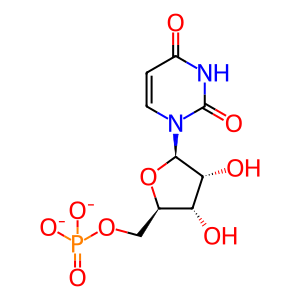
UMP [cytosol]
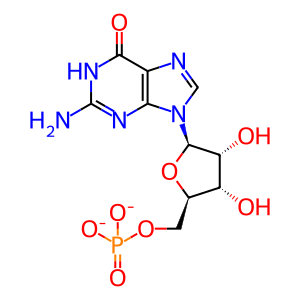
GMP [cytosol]
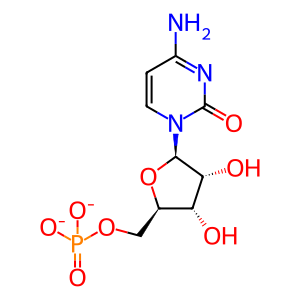
CMP [cytosol]
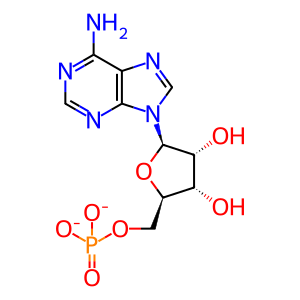
AMP [cytosol]
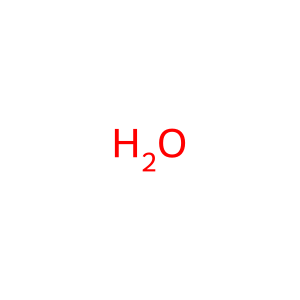
H2O [cytosol]
Reactome.org reaction link: R-HSA-430028
======
Reaction input - small molecules:
water
Reaction output - small molecules:
uridine 5'-monophosphate(2-)
guanosine 5'-monophosphate(2-)
cytidine 5'-monophosphate(2-)
adenosine 5'-monophosphate(2-)
Reactome.org link: R-HSA-430028