Reaction: SIRT6 deacetylates RBBP8
- in pathway: Processing of DNA double-strand break ends
RBBP8 (CtIP) is constitutively acetylated in the absence of DNA damage on lysine residues K432, K526 and K604, and perhaps other lysines. DNA damage, through an unknown mechanism, triggers deacetylation of RBBP8 by SIRT6 protein lysine deacetylase. SIRT6-mediated deacetylation of RBBP8 is necessary for RBBP8-promoted resection of DNA double-strand breaks (DSBs) (Kaidi et al. 2010).
Reaction - small molecule participants:
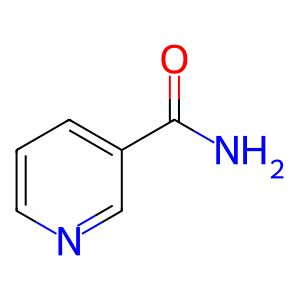
NAM [nucleoplasm]
2'-O-acetyl-ADP-ribose [nucleoplasm]
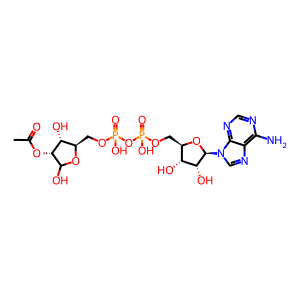
NAD+ [nucleoplasm]
Reactome.org reaction link: R-HSA-5685953
======
Reaction input - small molecules:
NAD(1-)
Reaction output - small molecules:
nicotinamide
2''-O-acetyl-ADP-D-ribose
Reactome.org link: R-HSA-5685953