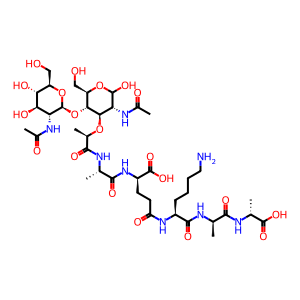
Reaction: PGLYRP3,4 binds bacterial peptidoglycan
PGRPs are thought to kill bacteria by interacting with cell wall peptidoglycan and by inducing lethal stress response in bacteria, as opposed to the hydrolysis of peptidoglycan or permeabilizing bacterial membranes seen with other antibacterial peptides (Lu X et al. 2006; Wang M et al. 2007; Cho S et al. 2007; Kashyap DR et al. 2011, 2014). In Gram‑positive bacteria, including B. subtilis, PGLYRP1, 3 & 4 were found to enter the bacterial cell wall at the site of daughter cell separation during cell division and to bind to cell wall peptidoglycan in the vicinity of the cell membrane (Kashyap DR et al. 2011). Binding of PGLYRP3 to peptidoglycan induces a structural change in PGLYRP3 that locks peptidoglycan in the protein's bindings groove (Guan R et al. 2006). However, this binding did not inhibit the extracellular transglycosylation or transpeptidation steps in peptidoglycan synthesis (Kashyap DR et al. 2011), which are well-known targets for bactericidal antibiotics. Instead, this interaction of PGRP proteins with peptidoglycan activated the B. subtilis envelope stress response two‑component system CssR‑CssS that detects and disposes of misfolded proteins exported out of bacterial cells. This activation resulted in bacterial membrane depolarization, cessation of intracellular peptidoglycan, protein, RNA, and DNA synthesis and production of toxic hydroxyl radicals, which were responsible for bacterial death (Kashyap DR et al. 2011). In Gram‑negative bacteria (E. coli), PGRPs were found to bind the bacterial outer membrane activating the functionally homologous CpxA‑CpxR two‑component system (Kashyap DR et al. 2011). Genome expression arrays, qRT‑PCR, and biochemical tests showed that PGLYRP3 & 4 kill both E. coli and B. subtilis by inducing oxidative, thiol, and metal stress (Kashyap DR et al. 2014).