Reaction: PGLYRP2 binds bacterial peptidoglycan
- in pathway: Antimicrobial peptides
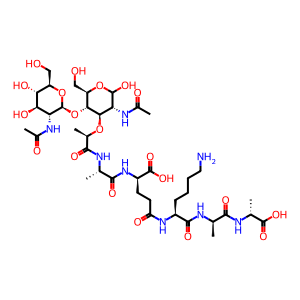
Peptidoglycan recognition proteins (PGRPs or PGLYRPs) are innate immunity molecules that contain a conserved peptidoglycan‑binding type 2 amidase domain that is homologous to bacteriophage and bacterial type 2 amidases (Kang D et al. 1998; Liu C et al. 2001; Royet J and Dziarski R 2007; Royet J et al. 2011; Dziarski R et al. 2016). Mammals have a family of four PGRPs (PGLYRP1, 2, 3 & 4) that are differentially expressed in a cell‑type‑ or tissue‑specific manner. PGLYRP2 (also known as PGRP-L) is constitutively expressed in the liver, from which it is secreted into blood as non-disulfide‑linked dimers (Liu C et al. 2001; Zhang Y et al. 2005; De Pauw P et al. 1995; Hoijer MA et al. 1996). PGLYRP2 expression can be also induced in the skin and intestine upon exposure to bacteria or pro-inflammatory cytokines (Wang H et al. 2005; Li X et al. 2006). The constitutive expression of PGLYRP2 in the liver and induced expression in epithelial cells is regulated by different transcription factors, the binding sequences for which are located in different regions of the pglyrp2 promoter (Li X et al. 2006). PGRP2 binds to bacterial cell wall peptidoglycan and functions as N‑acetylmuramoyl‑L‑alanine amidase that hydrolyzes the amide bond between the MurNAc and L‑alanine in peptidoglycan (Wang ZM et al. 2003; Zhang Y et al. 2005). The minimal peptidoglycan fragment hydrolyzed by PGLYRP2 is MurNAc-tripeptide (Wang ZM et al. 2003). PGLYRP2 has a conserved Zn(2+)‑binding site in the peptidoglycan‑binding groove, which is also present in bacteriophage type 2 amidases, and PGLYRP2 requires Zn(2+) for its amidase activity (Wang ZM et al. 2003). The amidase activity of mammalian PGLYRP2 is though to eliminate the pro‑inflammatory peptidoglycan and, therefore, prevent over‑activation of the immune system and excessive inflammation (Hoijer MA et al. 1997; Royet J and Dziarski R 2007). In addition to its amidase activity, PGLYRP2 also has antibacterial activity against both Gram-positive and Gram-negative bacteria and Chlamydia (Bobrovsky P et al. 2016), similar to PGLYRP1, PGLYRP3, and PGLYRP4 (Lu X et al. 2006; Wang M et al. 2007).
Reaction - small molecule participants:
GlcNac-(1-->4)MurNAc-L-Ala-gamma-D-Glu-L-Lys-(D-Ala)2 [peptidoglycan-based cell wall]
Reactome.org reaction link: R-HSA-6799981
======
Reaction input - small molecules:
beta-GlcNAc-(1->4)-MurNAc-L-Ala-gamma-D-Glu-L-Lys-(D-Ala)2
Reaction output - small molecules:
Reactome.org link: R-HSA-6799981