Reaction: DHODH:FMN oxidises (S)-DHO to orotate
- in pathway: Pyrimidine biosynthesis
Dihydroorotate dehydrogenase catalyzes the oxidation of dihydroorotate to orotate (orotic acid). The enzyme is located in the inner mitochondrial membrane oriented so that cytosolic dihydroorotate molecules have access to it, and orotate is released into the cytosol. The reducing equivalents generated by the reaction are transferred by ubiquinone (Coenzyme Q) to the electron transport chain within the inner mitochondrial membrane. There is no evidence for the involvement of NAD+ as an acceptor of reducing equivalents in this reaction in mammalian cells (Jones 1980). In contrast, the reaction catalyzed by the enzyme isolated from the anaerobic bacterium Clostridium oroticum requires NAD+ as a cofactor but also proceeds strongly in the direction of dihydroorotate synthesis consistent with the greater electronegativity of NAD+ (Lieberman and Kornberg 1953).
Early studies of purified rat liver enzyme by Forman and Kennedy suggested the presence of flavin mononucleotide and iron-sulfur complexes as cofactors. More recent work by Bader, Beuneu, and their colleagues with recombinant human protein expressed in insect cells has confirmed the presence of flavin mononucleotide, at a stoichiometry of one molecule per molecule of apoenzyme but suggests that iron-sulfur complexes, if indeed they are involved in the oxidation and electron transport process, are not an integral part of the dihydroorotate dehydrogenase holoenzyme.
Reaction - small molecule participants:
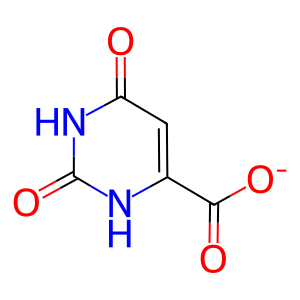
ORO [cytosol]
QH2 [mitochondrial inner membrane]
CoQ [mitochondrial inner membrane]
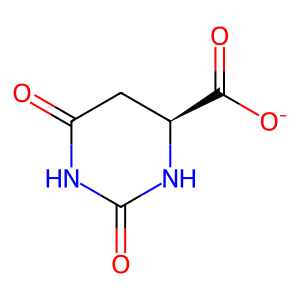
(S)-DHO [cytosol]
Reactome.org reaction link: R-HSA-73569
======
Reaction input - small molecules:
coenzyme Q10
(S)-dihydroorotate
Reaction output - small molecules:
orotate
ubiquinol
Reactome.org link: R-HSA-73569