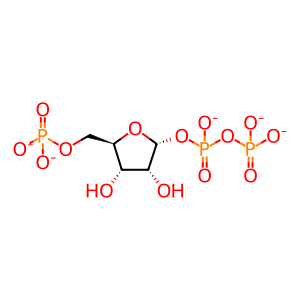
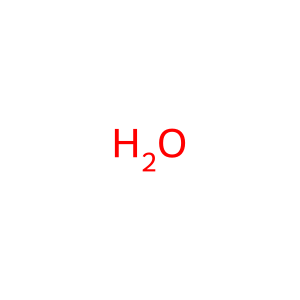
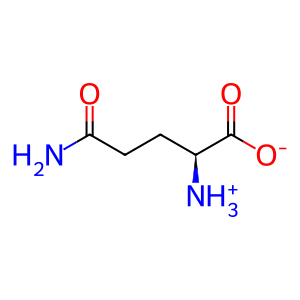
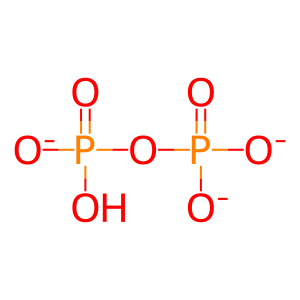
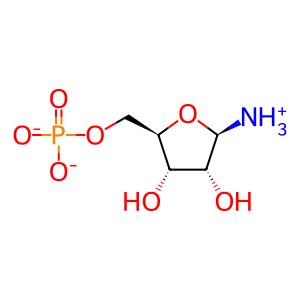
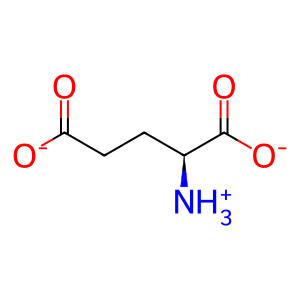
Reaction: 5-phospho-alpha-D-ribose 1-diphosphate (PRPP) + H2O + L-glutamine <=> 5-phosphoribosylamine + L-glutamate +pyrophosphate
- in pathway: Purine ribonucleoside monophosphate biosynthesis
Cytosolic PPAT (phosphoribosyl pyrophosphate amidotransferase) catalyzes the reaction of 5-phospho-alpha-D-ribose 1-diphosphate (PRPP), water, and L-glutamine to form 5-phosphoribosylamine, L-glutamate, and pyrophosphate. This event is the committed step in de novo purine synthesis. The reaction itself is reversible, but it is pulled strongly in the direction of 5'-phosphoribosylamine synthesis by the irreversible hydrolysis of the pyrophosphate that is also formed in the reaction. Fluoresence microscopy studies of cultured human cells have shown that PPAT is cytosolic and suggest that it may co-localize with other enzymes of de novo IMP biosynthesis under some metabolic conditions (An et al. 2008). The PPAT enzyme is inferred to be an iron-sulfur protein, like its well-characterized B. subtilis homologue, because incubation of purified enzyme with molecular oxygen or chelating agents inactivates it, and activity can be restored by incubation with ferrous iron and inorganic sulfide. The stoichiometry of the iron-sulfur moiety and its role in enzyme activity remain unknown (Itakura and Holmes 1979). The fully active form of the enzyme is a dimer, which can associate further to form a tetramer with sharply reduced activity (Holmes et al. 1973b; Iwahana et al. 1993). Interaction of the enzyme with inosine 5'-monophosphate (IMP), guanosine 5'-monophosphate (GMP), and adenosine 5'-monophosphate (AMP), end products of de novo purine biosynthesis, favors tetramer formation, while interaction with 5-phospho-alpha-D-ribose 1-diphosphate (PRPP), a required substrate, favors formation of the active dimer. Kinetic studies suggest that the enzyme's binding site for GMP and IMP is separate from its AMP binding site (Holmes et al. 1973a).
Reaction - small molecule participants:
PPi [cytosol]
PRA [cytosol]
L-Glu [cytosol]
PRPP [cytosol]
H2O [cytosol]
L-Gln [cytosol]
Reactome.org reaction link: R-HSA-73815
======
Reaction input - small molecules:
5-O-phosphonato-alpha-D-ribofuranosyl diphosphate(5-)
water
L-glutamine zwitterion
Reaction output - small molecules:
diphosphate(3-)
5-phospho-beta-D-ribosylaminium(1-)
L-glutamate(1-)
Reactome.org link: R-HSA-73815