Reaction: RET tyrosine phosphorylation
- in pathway: RET signaling
RET undergoes trans-autophosphorylation on specific tyrosine (Y) residues. The short and medium-length isoforms of RET contain 16 tyrosine residues; 10 in the kinase domain, 2 in the juxtamembrane domain, 1 in the kinase insert and 3 in the carboxy-terminal tail. The long RET isoform has 2 additional tyrosines in the carboxy-terminal tail. Phosphorylation of Y905 stabilizes the active conformation of the kinase and facilitates the autophosphorylation of Y residues mainly located in the C-terminal tail (Iwashita et al. 1996, Kawamoto et 2004). Y905, 1015, 1062 and 1096 are binding sites for GRB7/GRB10, phospholipase Cgamma (PLCG1), SHC1 and GRB2, respectively (Ichihara et al. 2004, Murakumo et al. 2006). Y1096 is present only in the long isoform. Phosphorylated Y981 is reported to bind SRC (Encinas et al. 2004).
RET can activate various signaling pathways including RAS-RAF-ERK (van Weering et al. 1995, Ohiwa et al. 1997, van Weering & Bos 1997, Trupp et al. 1999, Hayashi et al. 2000), phosphatidylinositol 3-kinase (PI3K)/AKT (Murakami et al. 1999a, Murakami et al. 1999b, Trupp et al. 1999, Soler et al. 1999, Segouffin-Cariou & Billaud 2000, Hayashi et al. 2000), p38 mitogen-activated protein kinase (MAPK) (Worby et al. 1996, Feng et al. 1999) and c-Jun N-terminal kinase (JNK) pathways (Xing et al. 1998, Chiariello et al. 1998). All these pathways are activated mainly through Y1062 (Hayashi et al. 2000). Point mutations at Y1062 result in a severe loss-of-function phenotype (Ibáñez 2013). SHC1 further associates with GRB2 and GAB1/GAB2, all of which become tyrosine phosphorylated. Tyrosine-phosphorylated GAB1/2 associates with the p85 subunit of PI3K, resulting in PI3K and AKT activation (Murakami et al. 1999b, Hayashi et al. 2000, Besset et al. 2000). GRB2-GAB1/2 can also assemble directly onto phosphorylated Y1096, an alternative route to PI3K activation (Besset et al. 2000). SHC1 can also form a complex with GRB2:SOS leading to activation of the RAS-RAF-ERK pathway (Hayashi et al. 2000). However, mutation of Y1062 did not completely abolish activation of the RAS-RAF-ERK and PI3K-AKT pathways suggesting alternative signaling pathways (Ichihara et al. 2004). The adaptor protein FRS2 can bind phosphorylated Y1062 (Kurokawa et al. 2001, Melillo et al. 2001), competing with SHC1 (Lundgren et al. 2006). Differential signaling may be mediated by different compartments in the plasma membrane, as RET has been shown to interact with FRS2 in lipid rafts, but with SHC1 outside lipid rafts (Paratcha et al. 2001).
Many other proteins have been shown to bind and/or become activated via Y1062. Docking protein 1 (DOK1), 2, 4, 5, and 6 adaptor proteins all interact with phosphorylated Y1062 (Grimm et al. 2001; Crowder et al. 2004; Kurotsuchi et al. 2010). Other suggested RET interactors include Mitogen-activated protein kinase 7 (MAPK7, BMK1) (Hayashi et al. 2001), SH3 and multiple ankyrin repeat domains protein 3 (SHANK3) (Schuetz et al. 2004), Insulin receptor substrate-2 (IRS2) (Hennige et al. 2000), SHC-transforming protein 3 (SHC3) (Pelicci et al. 2002), Protein kinase C alpha (PKCA) (Andreozzi et al. 2003) and PDZ and LIM domain protein 7 (Enigma, PDLIM7) (Durick et al. 1996). PDLIM7 and SHANK3 bind Y1062 regardless of its phosphorylation state.
Rap1GAP can bind phosphorylated Y981 to suppress GDNF-induced activation of ERK and neurite outgrowth (Jiao et al. 2011).
Tyrosine-protein phosphatase non-receptor type 11 (PTPN11, SHP2) binds to phosphorylated Y687 and components of the Y1062 associated signaling complex, contributing to activation of PI3K/AKT and promoting survival and neurite outgrowth in primary neurons (Besset et al. 2000, Perrinjaquet et al. 2010).
It is unclear how RET activates the p38MAPK, JNK, and ERK5 signaling pathways (Ichihara et al. 2004). To simplify the representation of RET signaling, all RET tyrosines known to be involved in signalling are phosphorylated in this event.
RET can activate various signaling pathways including RAS-RAF-ERK (van Weering et al. 1995, Ohiwa et al. 1997, van Weering & Bos 1997, Trupp et al. 1999, Hayashi et al. 2000), phosphatidylinositol 3-kinase (PI3K)/AKT (Murakami et al. 1999a, Murakami et al. 1999b, Trupp et al. 1999, Soler et al. 1999, Segouffin-Cariou & Billaud 2000, Hayashi et al. 2000), p38 mitogen-activated protein kinase (MAPK) (Worby et al. 1996, Feng et al. 1999) and c-Jun N-terminal kinase (JNK) pathways (Xing et al. 1998, Chiariello et al. 1998). All these pathways are activated mainly through Y1062 (Hayashi et al. 2000). Point mutations at Y1062 result in a severe loss-of-function phenotype (Ibáñez 2013). SHC1 further associates with GRB2 and GAB1/GAB2, all of which become tyrosine phosphorylated. Tyrosine-phosphorylated GAB1/2 associates with the p85 subunit of PI3K, resulting in PI3K and AKT activation (Murakami et al. 1999b, Hayashi et al. 2000, Besset et al. 2000). GRB2-GAB1/2 can also assemble directly onto phosphorylated Y1096, an alternative route to PI3K activation (Besset et al. 2000). SHC1 can also form a complex with GRB2:SOS leading to activation of the RAS-RAF-ERK pathway (Hayashi et al. 2000). However, mutation of Y1062 did not completely abolish activation of the RAS-RAF-ERK and PI3K-AKT pathways suggesting alternative signaling pathways (Ichihara et al. 2004). The adaptor protein FRS2 can bind phosphorylated Y1062 (Kurokawa et al. 2001, Melillo et al. 2001), competing with SHC1 (Lundgren et al. 2006). Differential signaling may be mediated by different compartments in the plasma membrane, as RET has been shown to interact with FRS2 in lipid rafts, but with SHC1 outside lipid rafts (Paratcha et al. 2001).
Many other proteins have been shown to bind and/or become activated via Y1062. Docking protein 1 (DOK1), 2, 4, 5, and 6 adaptor proteins all interact with phosphorylated Y1062 (Grimm et al. 2001; Crowder et al. 2004; Kurotsuchi et al. 2010). Other suggested RET interactors include Mitogen-activated protein kinase 7 (MAPK7, BMK1) (Hayashi et al. 2001), SH3 and multiple ankyrin repeat domains protein 3 (SHANK3) (Schuetz et al. 2004), Insulin receptor substrate-2 (IRS2) (Hennige et al. 2000), SHC-transforming protein 3 (SHC3) (Pelicci et al. 2002), Protein kinase C alpha (PKCA) (Andreozzi et al. 2003) and PDZ and LIM domain protein 7 (Enigma, PDLIM7) (Durick et al. 1996). PDLIM7 and SHANK3 bind Y1062 regardless of its phosphorylation state.
Rap1GAP can bind phosphorylated Y981 to suppress GDNF-induced activation of ERK and neurite outgrowth (Jiao et al. 2011).
Tyrosine-protein phosphatase non-receptor type 11 (PTPN11, SHP2) binds to phosphorylated Y687 and components of the Y1062 associated signaling complex, contributing to activation of PI3K/AKT and promoting survival and neurite outgrowth in primary neurons (Besset et al. 2000, Perrinjaquet et al. 2010).
It is unclear how RET activates the p38MAPK, JNK, and ERK5 signaling pathways (Ichihara et al. 2004). To simplify the representation of RET signaling, all RET tyrosines known to be involved in signalling are phosphorylated in this event.
Reaction - small molecule participants:
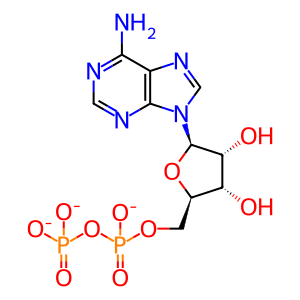
ADP [cytosol]
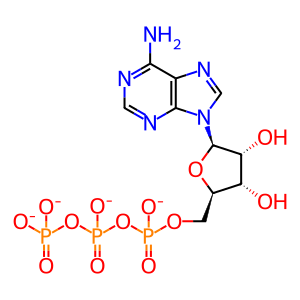
ATP [cytosol]
Reactome.org reaction link: R-HSA-8853792
======
Reaction input - small molecules:
ATP(4-)
Reaction output - small molecules:
ADP(3-)
Reactome.org link: R-HSA-8853792