Reaction: LYZ hydrolyzes peptidoglycans in the bacterial cell wall
- in pathway: Antimicrobial peptides
Lysozyme (LYZ), also known as 1,4-beta-N-acetylmuramidase C, is a host hydrolytic enzyme with muramidase activity that hydrolyzes (1->4)-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in the bacterial cell wall peptidoglycan (Schindler M et al. 1977; Surna A et al. 2009). LYZ acts against both gram-positive and gram-negative bacteria such as Peptostreptococcus micros, Eubacterium nodatum, Eikenella corrodens, Fusobacterium periodontium and Campylobacter rectus (Surna A et al. 2009; Tenovuo J 2002). The muramidase activity of LYZ is thought to be more effective against Gram-positive bacteria with their peptidoglycan layer exposed to the extracellular milieu. The detailed mechanism by which LYZ hydrolyses its substrate is described for hen egg-white lysozyme (HEWL) (Blake CC et al. 1967; Vocadlo DJ et al. 2001).
Many pathogens such as Streptococcus pneumoniae have evolved lysozyme resistance to prevent peptidoglycan hydrolysis. The primary mechanism for lysozyme resistance in both Gram-positive and Gram-negative organisms appears to be direct modification of peptidoglycan; however, modification of other cell wall-linked components, such as teichoic acid, may also contribute to resistance (Amano K & Williams JC 1983; Bera A et al. 2005; Pushkaran AC et al. 2015).
LYZ is found in many human secretions such as tears, milk, mucus and saliva (Surna A et al. 2009; Minami J et al. 2015; Sahin O et al. 2016; Masschalck B & Michiels CW. 2003).
Reaction - small molecule participants:
betaGlcNAc [peptidoglycan-based cell wall]
H2O [extracellular region]
Reactome.org reaction link: R-HSA-8862320
======
Reaction input - small molecules:
water
Reaction output - small molecules:
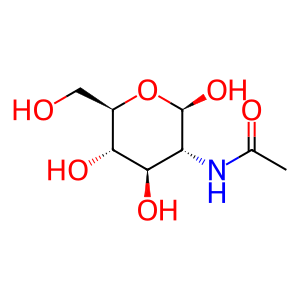
N-acetyl-beta-D-glucosamine
Reactome.org link: R-HSA-8862320