Reaction: PARPs transfer ADP-D-ribose to proteins (poly(ADP-ribosyl)ation)
- in pathway: Nicotinamide salvaging
Poly (ADP-ribose) polymerases (PARPs) catalyse the poly(ADP-ribosyl)ation posttranslational modification of proteins. At least 18 human members share homology with the catalytic domain of the founding member, PARP1. PARPs cleave the glycosidic bond of NAD+ between nicotinamide (NAM) and ribose followed by the covalent modification of mainly glutamate residues on acceptor proteins with an ADP-ribosyl unit, with subsequent ADP-ribosyl unit additions linked by glycosidic ribose-ribose bonds. NAM can be utilised in the NAD+ regeneration process. Poly(ADP-ribosyl)ation is important in many biological processess including DNA repair, regulation of chromosome structure, transcriptional regulation, mitosis and apoptosis. PARPs can localise to either the cytosol or the nucleus. The cytosolic PARPs described here are PARP9, PARP10 and PARP16 (Yan et al. 2013, Yu et al. 2005, Di Paolo et al. 2012). PARP4, PARP6, PARP8 and PARP14 may also be located in the cytosol with the same functionality.
Reaction - small molecule participants:
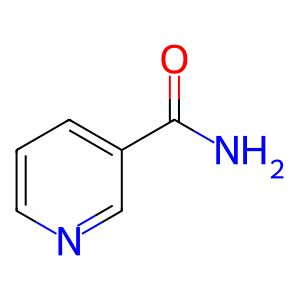
NAM [cytosol]
(ADP-D-ribosyl)(n+1)-acceptor [cytosol]
(ADP-D-ribosyl)(n)-acceptor [cytosol]
NAD+ [cytosol]
Reactome.org reaction link: R-HSA-8938073
======
Reaction input - small molecules:
(ADP-D-ribosyl)(n)-acceptor
NAD(1-)
Reaction output - small molecules:
nicotinamide
(ADP-D-ribosyl)(n+1)-acceptor
Reactome.org link: R-HSA-8938073