Reaction: RHOA GAPs stimulate RHOA GTPase activity
- in pathway: RHOA GTPase cycle
The following GTPase activating proteins (GAPs) were shown to bind RHOA and stimulate its GTPase activity, resulting in GTP to GDP hydrolysis and conversion of the active RHOA:GTP complex into the inactive RHOA:GDP complex (the high throughput study by Bagci et al. 2020 looked at preferential binding of GAPs to active RHOA mutant compared to wild type RHOA but did not examine activation of GTPase activity):
ARHGAP5 (Burbelo et al. 1995; Kusama et al. 2006; Amin et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP6 (Prakash et al. 2000; Müller et al. 2020)
ARHGAP8 (Lua and Low 2004; Müller et al. 2020)
ARHGAP10 (Shibata et al. 2001; Müller et al. 2020)
ARHGAP11A (Lawson et al. 2016; Müller et al. 2020)
ARHGAP11B (Müller et al. 2020)
ARHGAP19 (David et al. 2014; Müller et al. 2020)
ARHGAP20 (Müller et al. 2020)
ARHGAP21 (Sousa et al. 2005; Lazarini et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP23 (Müller et al. 2020)
ARHGAP26 (Hildebrand et al. 1996; Sheffield et al. 1999; Amin et al. 2016; Müller et al. 2020)
ARHGAP28 (Yeung et al. 2014; Müller et al. 2020)
ARHGAP29 (Saras et al. 1997; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP30 (Naji et al. 2011; Müller et al. 2020)
ARHGAP40 (Müller et al. 2020)
DLC1 (Healy et al. 2008; Amin et al. 2016; Müller et al. 2020)
GMIP (Aresta et al. 2002; Müller et al. 2020)
HMHA1 (de Kreuk et al. 2013)
MYO9A (Handa et al. 2013; Omelchenko and Hall 2012; Müller et al. 2020; supported by Bagci et al. 2020)
MYO9B (Post et al. 1998; Graf et al. 2000; Kong et al. 2015; Müller et al. 2020; supported by Bagci et al. 2020)
TAGAP (Bauer et al. 2005; Müller et al. 2020)
The following GAPs were shown to bind RHOA and stimulate its GTPase activity in some but not all studies and are annotated as candidate RHOA GAPs:
ABR (Bagci et al. 2020: binding to active RHOA; Chuang et al. 1995, Amin et al. 2016, Müller et al. 2020: no activation of RHOA GTPase activity)
ARAP1 (Miura et al. 2002: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARAP2 (Müller et al. 2020: activation of RHOA GTPase activity; Yoon et al. 2006: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARAP3 (Krugmann et al. 2002: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP1 (Zhang and Zheng 1998, Amin et al. 2016, Müller et al. 2020: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Yang et al. 2006, Li et al. 2009: no activation of RHOA GTPase activity)
ARHGAP4 (Vogt et al. 2007: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP9 (Furukawa et al. 2001: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP18 (Maeda et al. 2011: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP22 (Müller et al. 2020: activation of RHOA GTPase activity; Mori et al. 2014: no activation of RHOA GTPase activity)
ARHGAP24 (Su et al. 2004: activation of RHOA GTPase activity; Lavelin and Geiger 2005, Ohta et al. 2006, Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP31 (Müller et al. 2020: activation of RHOA GTPase activity; Tcherkezian et al. 2006: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP32 (Nakazawa et al. 2003: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP35 (Levay et al. 2009, Amin et al. 2016; Müller et al. 2020: activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP39 (Bagci et al. 2020: binding to active RHOA; Lundstrom et al. 2004, Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP42 (Bai et al. 2013, Luo et al. 2017, Bai et al. 2017: activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP44 (Raynaud et al. 2014: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
BCR (Zhang and Zheng 1998: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Chuang et al. 1995, Müller et al. 2020: no activation of RHOA GTPase activity),
DEPDC1B (Bagci et al. 2020: binding to active RHOA; Wu et al. 2015, Müller et al. 2020: no activation of RHOA GTPase activity)
FAM13A (Li et al. 2015: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
OPHN1 (Elvers et al. 2012, Amin et al. 2016: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
PIK3R1 (Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
PIK3R2 (Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
RACGAP1 (Toure et al. 1998; Amin et al. 2016: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
SRGAP1 (Wong et al. 2001: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
STARD8 (Kawai et al. 2007, Müller et al. 2020: activation of RHOA GTPase activity; Amin et al. 2016: no activation of RHOA GTPase activity)
STARD13 (Ching et al. 2003, Leung et al. 2005, Müller et al. 2020: activation of RHOA GTPase activity; Amin et al. 2016: no activation of RHOA GTPase activity)
The following GAPs do not act on RHOA or were shown to not bind to active RHOA without testing for activation of RHOA GTPase activity:
ARHGAP12 (Müller et al. 2020; Bagci et al. 2020: no binding to RHOA)
ARHGAP15 (Seoh et al. 2003; Müller et al. 2020)
ARHGAP17 (Richnau and Aspenstrom 2001; Amin et al. 2016; Müller et al. 2020; Bagci et al. 2020: no binding to RHOA)
ARHGAP25 (Csepanyi Komi et al. 2012; Müller et al. 2020)
ARHGAP27 (Sakakibara et al. 2004; Müller et al. 2020)
ARHGAP33 (Liu et al. 2006; Müller et al. 2020)
ARHGAP36 (Müller et al. 2020)
ARHGAP45 (Müller et al. 2020)
CHN1 (Ahmed et al. 1994; Müller et al. 2020)
CHN2 (Caloca et al. 2003; Müller et al. 2020)
DEPDC1 (Müller et al. 2020)
FAM13B (Müller et al. 2020)
INPP5B (Müller et al. 2020)
OCRL (Lichter Konecki et al. 2006; Erdmann et al. 2007; Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
RALBP1 (Jullien Flores et al. 1995; Müller et al. 2020)
SH3BP1 (Müller et al. 2020)
SRGAP2 (Mason et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
SRGAP3 (Endris et al. 2002; Müller et al. 2020)
SYDE1 (Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
SYDE2 (Müller et al. 2020)
ARHGAP5 (Burbelo et al. 1995; Kusama et al. 2006; Amin et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP6 (Prakash et al. 2000; Müller et al. 2020)
ARHGAP8 (Lua and Low 2004; Müller et al. 2020)
ARHGAP10 (Shibata et al. 2001; Müller et al. 2020)
ARHGAP11A (Lawson et al. 2016; Müller et al. 2020)
ARHGAP11B (Müller et al. 2020)
ARHGAP19 (David et al. 2014; Müller et al. 2020)
ARHGAP20 (Müller et al. 2020)
ARHGAP21 (Sousa et al. 2005; Lazarini et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP23 (Müller et al. 2020)
ARHGAP26 (Hildebrand et al. 1996; Sheffield et al. 1999; Amin et al. 2016; Müller et al. 2020)
ARHGAP28 (Yeung et al. 2014; Müller et al. 2020)
ARHGAP29 (Saras et al. 1997; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP30 (Naji et al. 2011; Müller et al. 2020)
ARHGAP40 (Müller et al. 2020)
DLC1 (Healy et al. 2008; Amin et al. 2016; Müller et al. 2020)
GMIP (Aresta et al. 2002; Müller et al. 2020)
HMHA1 (de Kreuk et al. 2013)
MYO9A (Handa et al. 2013; Omelchenko and Hall 2012; Müller et al. 2020; supported by Bagci et al. 2020)
MYO9B (Post et al. 1998; Graf et al. 2000; Kong et al. 2015; Müller et al. 2020; supported by Bagci et al. 2020)
TAGAP (Bauer et al. 2005; Müller et al. 2020)
The following GAPs were shown to bind RHOA and stimulate its GTPase activity in some but not all studies and are annotated as candidate RHOA GAPs:
ABR (Bagci et al. 2020: binding to active RHOA; Chuang et al. 1995, Amin et al. 2016, Müller et al. 2020: no activation of RHOA GTPase activity)
ARAP1 (Miura et al. 2002: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARAP2 (Müller et al. 2020: activation of RHOA GTPase activity; Yoon et al. 2006: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARAP3 (Krugmann et al. 2002: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP1 (Zhang and Zheng 1998, Amin et al. 2016, Müller et al. 2020: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Yang et al. 2006, Li et al. 2009: no activation of RHOA GTPase activity)
ARHGAP4 (Vogt et al. 2007: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP9 (Furukawa et al. 2001: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP18 (Maeda et al. 2011: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP22 (Müller et al. 2020: activation of RHOA GTPase activity; Mori et al. 2014: no activation of RHOA GTPase activity)
ARHGAP24 (Su et al. 2004: activation of RHOA GTPase activity; Lavelin and Geiger 2005, Ohta et al. 2006, Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP31 (Müller et al. 2020: activation of RHOA GTPase activity; Tcherkezian et al. 2006: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP32 (Nakazawa et al. 2003: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP35 (Levay et al. 2009, Amin et al. 2016; Müller et al. 2020: activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP39 (Bagci et al. 2020: binding to active RHOA; Lundstrom et al. 2004, Müller et al. 2020: no activation of RHOA GTPase activity)
ARHGAP42 (Bai et al. 2013, Luo et al. 2017, Bai et al. 2017: activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
ARHGAP44 (Raynaud et al. 2014: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
BCR (Zhang and Zheng 1998: activation of RHOA GTPase activity; Bagci et al. 2020: binding to active RHOA; Chuang et al. 1995, Müller et al. 2020: no activation of RHOA GTPase activity),
DEPDC1B (Bagci et al. 2020: binding to active RHOA; Wu et al. 2015, Müller et al. 2020: no activation of RHOA GTPase activity)
FAM13A (Li et al. 2015: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
OPHN1 (Elvers et al. 2012, Amin et al. 2016: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
PIK3R1 (Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
PIK3R2 (Bagci et al. 2020: binding to active RHOA; Müller et al. 2020: no activation of RHOA GTPase activity)
RACGAP1 (Toure et al. 1998; Amin et al. 2016: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity; Bagci et al. 2020: no binding to active RHOA)
SRGAP1 (Wong et al. 2001: activation of RHOA GTPase activity; Müller et al. 2020: no activation of RHOA GTPase activity)
STARD8 (Kawai et al. 2007, Müller et al. 2020: activation of RHOA GTPase activity; Amin et al. 2016: no activation of RHOA GTPase activity)
STARD13 (Ching et al. 2003, Leung et al. 2005, Müller et al. 2020: activation of RHOA GTPase activity; Amin et al. 2016: no activation of RHOA GTPase activity)
The following GAPs do not act on RHOA or were shown to not bind to active RHOA without testing for activation of RHOA GTPase activity:
ARHGAP12 (Müller et al. 2020; Bagci et al. 2020: no binding to RHOA)
ARHGAP15 (Seoh et al. 2003; Müller et al. 2020)
ARHGAP17 (Richnau and Aspenstrom 2001; Amin et al. 2016; Müller et al. 2020; Bagci et al. 2020: no binding to RHOA)
ARHGAP25 (Csepanyi Komi et al. 2012; Müller et al. 2020)
ARHGAP27 (Sakakibara et al. 2004; Müller et al. 2020)
ARHGAP33 (Liu et al. 2006; Müller et al. 2020)
ARHGAP36 (Müller et al. 2020)
ARHGAP45 (Müller et al. 2020)
CHN1 (Ahmed et al. 1994; Müller et al. 2020)
CHN2 (Caloca et al. 2003; Müller et al. 2020)
DEPDC1 (Müller et al. 2020)
FAM13B (Müller et al. 2020)
INPP5B (Müller et al. 2020)
OCRL (Lichter Konecki et al. 2006; Erdmann et al. 2007; Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
RALBP1 (Jullien Flores et al. 1995; Müller et al. 2020)
SH3BP1 (Müller et al. 2020)
SRGAP2 (Mason et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
SRGAP3 (Endris et al. 2002; Müller et al. 2020)
SYDE1 (Müller et al. 2020; Bagci et al. 2020: no binding to active RHOA)
SYDE2 (Müller et al. 2020)
Reaction - small molecule participants:
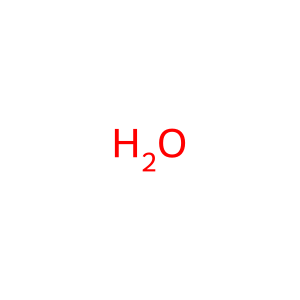
H2O [cytosol]
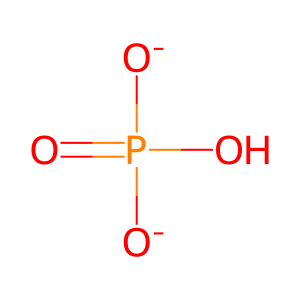
Pi [cytosol]
Reactome.org reaction link: R-HSA-8981637
======
Reaction input - small molecules:
water
Reaction output - small molecules:
hydrogenphosphate
Reactome.org link: R-HSA-8981637