Reaction: CDC42 GAPs stimulate CDC42 GTPase activity
- in pathway: CDC42 GTPase cycle
The following GTPase activating proteins (GAPs) were shown to bind CDC42 and stimulate its GTPase activity, resulting in GTP to GDP hydrolysis and conversion of the active CDC42:GTP complex into the inactive CDC42:GDP complex (the high throughput screen by Bagci et al. 2020 is cited as supporting evidence since it examined binding of GAPs to constitutively active CDC42 mutant but did not test for activation of CDC42 GTPase activity):
ARAP1 (Miura et al. 2002; Müller et al. 2020)
ARHGAP1 (Nassar et al. 1998; Amin et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP20 (Müller et al. 2020)
ARHGAP22 (Mori et al. 2014; Müller et al. 2020)
ARHGAP39 (Lundström et al. 2004; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP40 (Müller et al. 2020)
FAM13B (Müller et al. 2020)
HMHA1 (de Kreuk et al. 2013)
The following GAPs were shown to bind CDC42 and stimulate its GTPase activity in some but not all studies and are annotated as candidate CDC42 GAPs:
ABR (Chuang et al. 1995, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42 GAP)
ARAP2 (Müller et al. 2020: CDC42-directed GAP activity; Yoon et al. 2006: no CDC42-directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARAP3 (Krugmann et al. 2002, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP4 (Vogt et al. 2007: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP5 (Burbelo et al. 1995: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP9 (Furukawa et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP10 (also known as Graf2) (Shibata et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP11B (Müller et al. 2020: CDC42-directed GAP activity; Florio et al. 2015: no CDC42-directed GAP activity)
ARHGAP17 (Richnau and Aspenstrom 2001, Amin et al. 2016: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP21 (Sousa et al. 2005: CDC42 directed GAP activity; Bagci et al. 2020: binding to active CDC42; Lazarini et al. 2013, Müller et al. 2020: no CDC42 directed GAP activity)
ARHGAP24 (Lavelin and Geiger 2005, Ohta et al. 2006: CDC42 directed GAP activity; Su et al. 2004, Müller et al. 2020: no CDC42 directed GAP activity)
ARHGAP26 (Hildebrand 1996, Sheffield et al. 1999, Amin et al. 2016: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP27 (Sakakibara et al. 2004: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP29 (Saras et al. 1997: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP30 (Müller et al. 2020: CDC42-directed GAP activity; Naji et al. 2011: no CDC42-directed GAP activity)
ARHGAP31 (Tcherkezian et al. 2006, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP32 (Nakazawa et al. 2003: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP33 (Chiang et al. 2003, Liu et al. 2006: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP35 (Zhang et al. 1997, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP42 (Bai et al. 2013: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP44 (also known as RICH2) (Raynaud et al. 2014: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
BCR (Chuang et al. 1995: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
CHN1 (also known as ARHGAP2) (Ahmed et al. 1994: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
DEPDC1B (Bagci et al. 2020: binding to active CDC42; Wu et al. 2015, Müller et al. 2020: no CDC42 directed GAP activity)
DLC1 (Healy et al. 2008, Kim et al. 2008, Amin et al. 2016: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
GMIP (Müller et al. 2020: CDC42-directed GAP activity; Aresta et al. 2002: no CDC42-directed GAP activity)
MYO9B (Müller et al. 2020: CDC42-directed GAP activity; Post et al. 1998, Kong et al. 2015: no CDC42-directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
OPHN1 (Elvers et al. 2012, Amin et al. 2016: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
PIK3R1 (Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
PIK3R2 (Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
RACGAP1 (Touré et al. 1998, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
RALBP1 (Jullien Flores et al. 1995: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
SRGAP1 (Wong et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
SRGAP2 (Mason et al. 2011, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
SRGAP3 (Endris et al. 2002: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
STARD8 (Kawai et al. 2007, Müller et al. 2020: CDC42 directed GAP activity; Amin et al. 2016: no CDC42 directed GAP activity)
STARD13 (Leung et al. 2005, Ching et al. 2003: CDC42 directed GAP activity; Amin et al. 2016, Müller et al. 2020: no CDC42 directed GAP activity)
SYDE1 (Amado Azevedo et al. 2017: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
TAGAP (Bauer et al. 2005: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
The following GAPs do not act on CDC42 or were shown in the high throughput screen by Bagci et al. 2020 to not bind to constitutively active CDC42 mutant:
ARHGAP6 (Prakash et al. 2000; Müller et al. 2020)
ARHGAP8 (Lua and Low 2004; Müller et al. 2020)
ARHGAP11A (Lawson et al. 2016; Müller et al. 2020)
ARHGAP12 (Bagci et al. 2020; Müller et al. 2020)
ARHGAP15 (Seoh et al. 2003; Müller et al. 2020)
ARHGAP18 (Maeda et al. 2011; Müller et al. 2020)
ARHGAP19 (David et al. 2014; Müller et al. 2020)
ARHGAP23 (Müller et al. 2020)
ARHGAP25 (Csépányi Kömi et al. 2012; Müller et al. 2020)
ARHGAP28 (Yeung et al. 2014; Müller et al. 2020)
ARHGAP36 (Rack et al. 2014; Müller et al. 2020; ARHGAP36 was shown by Jelen et al. 2009 to lack motifs needed for the GAP activity and likely does not act like a GAP)
ARHGAP45 (Müller et al. 2020)
CHN2 (Caloca et al. 2003; Müller et al. 2020)
DEPDC1 (Müller et al. 2020)
FAM13A (Müller et al. 2020)
INPP5B (Müller et al. 2020)
MYO9A (Müller et al. 2020; Bagci et al. 2020)
OCRL (Erdmann et al. 2007; Lichter Konecki et al. 2006; Müller et al. 2020; Bagci et al. 2020)
SH3BP1 (Müller et al. 2020)
SYDE2 (Müller et al. 2020)
ARAP1 (Miura et al. 2002; Müller et al. 2020)
ARHGAP1 (Nassar et al. 1998; Amin et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP20 (Müller et al. 2020)
ARHGAP22 (Mori et al. 2014; Müller et al. 2020)
ARHGAP39 (Lundström et al. 2004; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGAP40 (Müller et al. 2020)
FAM13B (Müller et al. 2020)
HMHA1 (de Kreuk et al. 2013)
The following GAPs were shown to bind CDC42 and stimulate its GTPase activity in some but not all studies and are annotated as candidate CDC42 GAPs:
ABR (Chuang et al. 1995, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42 GAP)
ARAP2 (Müller et al. 2020: CDC42-directed GAP activity; Yoon et al. 2006: no CDC42-directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARAP3 (Krugmann et al. 2002, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP4 (Vogt et al. 2007: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP5 (Burbelo et al. 1995: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP9 (Furukawa et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP10 (also known as Graf2) (Shibata et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP11B (Müller et al. 2020: CDC42-directed GAP activity; Florio et al. 2015: no CDC42-directed GAP activity)
ARHGAP17 (Richnau and Aspenstrom 2001, Amin et al. 2016: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP21 (Sousa et al. 2005: CDC42 directed GAP activity; Bagci et al. 2020: binding to active CDC42; Lazarini et al. 2013, Müller et al. 2020: no CDC42 directed GAP activity)
ARHGAP24 (Lavelin and Geiger 2005, Ohta et al. 2006: CDC42 directed GAP activity; Su et al. 2004, Müller et al. 2020: no CDC42 directed GAP activity)
ARHGAP26 (Hildebrand 1996, Sheffield et al. 1999, Amin et al. 2016: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP27 (Sakakibara et al. 2004: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP29 (Saras et al. 1997: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP30 (Müller et al. 2020: CDC42-directed GAP activity; Naji et al. 2011: no CDC42-directed GAP activity)
ARHGAP31 (Tcherkezian et al. 2006, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP32 (Nakazawa et al. 2003: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP33 (Chiang et al. 2003, Liu et al. 2006: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
ARHGAP35 (Zhang et al. 1997, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP42 (Bai et al. 2013: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
ARHGAP44 (also known as RICH2) (Raynaud et al. 2014: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
BCR (Chuang et al. 1995: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
CHN1 (also known as ARHGAP2) (Ahmed et al. 1994: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
DEPDC1B (Bagci et al. 2020: binding to active CDC42; Wu et al. 2015, Müller et al. 2020: no CDC42 directed GAP activity)
DLC1 (Healy et al. 2008, Kim et al. 2008, Amin et al. 2016: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
GMIP (Müller et al. 2020: CDC42-directed GAP activity; Aresta et al. 2002: no CDC42-directed GAP activity)
MYO9B (Müller et al. 2020: CDC42-directed GAP activity; Post et al. 1998, Kong et al. 2015: no CDC42-directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
OPHN1 (Elvers et al. 2012, Amin et al. 2016: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
PIK3R1 (Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
PIK3R2 (Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
RACGAP1 (Touré et al. 1998, Amin et al. 2016: CDC42 directed GAP activity; Müller et al. 2020: no CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
RALBP1 (Jullien Flores et al. 1995: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
SRGAP1 (Wong et al. 2001: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
SRGAP2 (Mason et al. 2011, Müller et al. 2020: CDC42 directed GAP activity; Bagci et al. 2020: no binding to active CDC42)
SRGAP3 (Endris et al. 2002: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
STARD8 (Kawai et al. 2007, Müller et al. 2020: CDC42 directed GAP activity; Amin et al. 2016: no CDC42 directed GAP activity)
STARD13 (Leung et al. 2005, Ching et al. 2003: CDC42 directed GAP activity; Amin et al. 2016, Müller et al. 2020: no CDC42 directed GAP activity)
SYDE1 (Amado Azevedo et al. 2017: CDC42-directed GAP activity; Bagci et al. 2020: binding to active CDC42; Müller et al. 2020: no CDC42-directed GAP activity)
TAGAP (Bauer et al. 2005: CDC42-directed GAP activity; Müller et al. 2020: no CDC42-directed GAP activity)
The following GAPs do not act on CDC42 or were shown in the high throughput screen by Bagci et al. 2020 to not bind to constitutively active CDC42 mutant:
ARHGAP6 (Prakash et al. 2000; Müller et al. 2020)
ARHGAP8 (Lua and Low 2004; Müller et al. 2020)
ARHGAP11A (Lawson et al. 2016; Müller et al. 2020)
ARHGAP12 (Bagci et al. 2020; Müller et al. 2020)
ARHGAP15 (Seoh et al. 2003; Müller et al. 2020)
ARHGAP18 (Maeda et al. 2011; Müller et al. 2020)
ARHGAP19 (David et al. 2014; Müller et al. 2020)
ARHGAP23 (Müller et al. 2020)
ARHGAP25 (Csépányi Kömi et al. 2012; Müller et al. 2020)
ARHGAP28 (Yeung et al. 2014; Müller et al. 2020)
ARHGAP36 (Rack et al. 2014; Müller et al. 2020; ARHGAP36 was shown by Jelen et al. 2009 to lack motifs needed for the GAP activity and likely does not act like a GAP)
ARHGAP45 (Müller et al. 2020)
CHN2 (Caloca et al. 2003; Müller et al. 2020)
DEPDC1 (Müller et al. 2020)
FAM13A (Müller et al. 2020)
INPP5B (Müller et al. 2020)
MYO9A (Müller et al. 2020; Bagci et al. 2020)
OCRL (Erdmann et al. 2007; Lichter Konecki et al. 2006; Müller et al. 2020; Bagci et al. 2020)
SH3BP1 (Müller et al. 2020)
SYDE2 (Müller et al. 2020)
Reaction - small molecule participants:
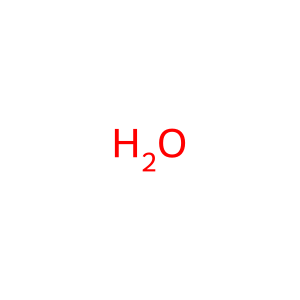
H2O [cytosol]
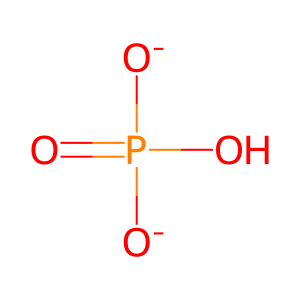
Pi [cytosol]
Reactome.org reaction link: R-HSA-9013161
======
Reaction input - small molecules:
water
Reaction output - small molecules:
hydrogenphosphate
Reactome.org link: R-HSA-9013161