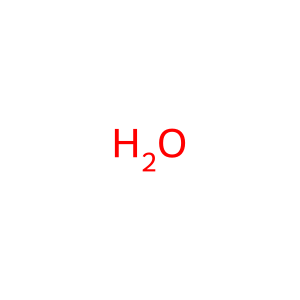
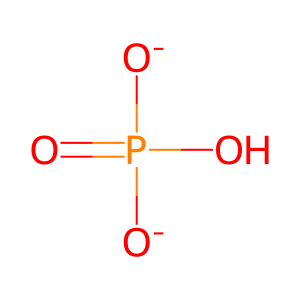
Reaction: RAC2 GAPs stimulate RAC2 GTPase activity
- in pathway: RAC2 GTPase cycle
The following GTPase -activating proteins (GAPs) were shown to bind RAC2 and stimulate its GTPase activity, resulting in GTP to GDP hydrolysis and conversion of the active RAC2:GTP complex into the inactive RAC2:GDP complex (the high throughput study by Bagci et al. 2020 is cited as supporting evidence, because it examined binding of GAPs to constitutively active RAC2 mutant but did not examine GAP-mediated activation of RAC2 GTPase activity):
ARHGAP1 (Amin et al. 2016; supported by Bagci et al. 2020)
ARHGAP26 (Amin et al. 2016)
ARHGAP35 (Amin et al. 2016; Bagci et al. 2020)
The following GAPs, annotated as candidate RAC2 GAPs, were shown to bind RAC2 and stimulate its GTPase activity in some but not all studies or were shown to bind to constitutively active RAC2 mutant (Bagci et al. 2020) but their ability to activate RAC2 GTPase activity has not been tested:
ABR (Chuang et al. 1995: RAC2 directed GAP activity; Amin et al. 2016: no RAC2 directed GAP activity; Bagci et al. 2020: no binding t active RAC2)
ARHGAP17 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
ARHGAP21 (Bagci et al. 2020: binding to active RAC2)
ARHGAP32 (Bagci et al. 2020: binding to active RAC2)
ARHGAP39 (Bagci et al. 2020: binding to active RAC2)
ARHGAP42 (Bagci et al. 2020: binding to active RAC2)
BCR (Chuang et al. 1995: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
DEPDC1B (Bagci et al. 2020)
OPHN1 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
PIK3R1 (Bagci et al. 2020: binding to active RAC2)
PIK3R2 (Bagci et al. 2020: binding to active RAC2)
RACGAP1 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
SYDE1 (Bagci et al. 2020: binding to active RAC2)
The following GAPs do not act on RAC2 or were shown by Bagci et al. 2020 to not bind to active RAC2:
ARAP2 (Bagci et al. 2020)
ARAP3 (Bagci et al. 2020)
ARHGAP5 (Bagci et al. 2020)
ARHGAP12 (Bagci et al. 2020)
ARHGAP29 (Bagci et al. 2020)
ARHGAP31 (Bagci et al. 2020)
DLC1 (Amin et al. 2016)
MYO9A (Bagci et al. 2020)
MYO9B (Bagci et al. 2020)
OCRL (Lichter Konecki et al. 2006; Erdmann et al. 2007; Bagci et al. 2020)
SRGAP2 (Bagci et al. 2020)
STARD8 (Amin et al. 2016)
STARD13 (Amin et al. 2016)
ARHGAP1 (Amin et al. 2016; supported by Bagci et al. 2020)
ARHGAP26 (Amin et al. 2016)
ARHGAP35 (Amin et al. 2016; Bagci et al. 2020)
The following GAPs, annotated as candidate RAC2 GAPs, were shown to bind RAC2 and stimulate its GTPase activity in some but not all studies or were shown to bind to constitutively active RAC2 mutant (Bagci et al. 2020) but their ability to activate RAC2 GTPase activity has not been tested:
ABR (Chuang et al. 1995: RAC2 directed GAP activity; Amin et al. 2016: no RAC2 directed GAP activity; Bagci et al. 2020: no binding t active RAC2)
ARHGAP17 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
ARHGAP21 (Bagci et al. 2020: binding to active RAC2)
ARHGAP32 (Bagci et al. 2020: binding to active RAC2)
ARHGAP39 (Bagci et al. 2020: binding to active RAC2)
ARHGAP42 (Bagci et al. 2020: binding to active RAC2)
BCR (Chuang et al. 1995: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
DEPDC1B (Bagci et al. 2020)
OPHN1 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
PIK3R1 (Bagci et al. 2020: binding to active RAC2)
PIK3R2 (Bagci et al. 2020: binding to active RAC2)
RACGAP1 (Amin et al. 2016: RAC2 directed GAP activity; Bagci et al. 2020: no binding to active RAC2)
SYDE1 (Bagci et al. 2020: binding to active RAC2)
The following GAPs do not act on RAC2 or were shown by Bagci et al. 2020 to not bind to active RAC2:
ARAP2 (Bagci et al. 2020)
ARAP3 (Bagci et al. 2020)
ARHGAP5 (Bagci et al. 2020)
ARHGAP12 (Bagci et al. 2020)
ARHGAP29 (Bagci et al. 2020)
ARHGAP31 (Bagci et al. 2020)
DLC1 (Amin et al. 2016)
MYO9A (Bagci et al. 2020)
MYO9B (Bagci et al. 2020)
OCRL (Lichter Konecki et al. 2006; Erdmann et al. 2007; Bagci et al. 2020)
SRGAP2 (Bagci et al. 2020)
STARD8 (Amin et al. 2016)
STARD13 (Amin et al. 2016)
Reaction - small molecule participants:
Pi [cytosol]
H2O [cytosol]
Reactome.org reaction link: R-HSA-9014295
======
Reaction input - small molecules:
water
Reaction output - small molecules:
hydrogenphosphate
Reactome.org link: R-HSA-9014295