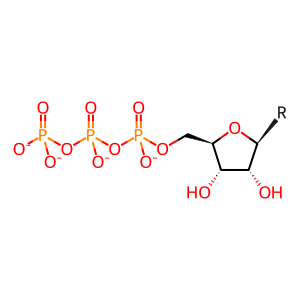
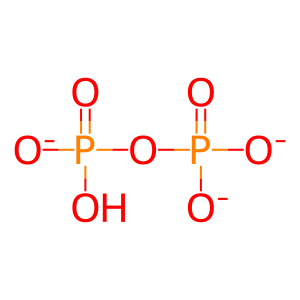
Reaction: nsp12 misincorporates a nucleotide in nascent RNA minus strand
- in pathway: Replication of the SARS-CoV-1 genome
In the presence of functional nsp14, which acts as a 3'-to-5' exonuclease, the mutation rate during human SARS coronavirus 1 (SARS-CoV-1) replication is 9 x 10^-7 (9E-7) per nucleotide per replication cycle or 2.2 x 10^-5 (2.2E-5) non-redundant substitutions per nucleotide, which translates into 2-3 nucleotide substitutions for each replicated SARS-CoV-1 genome. When nsp14 is defective, the mutation rate during SARS-CoV-1 replication increases to 1.2 x 10^-5 (1.2E-5) mutations per nucleotide per replication cycle or 3.34 x 10^-4 (3.34E-4) non-redundant substitutions per nucleotide, which translates into 12-23 nucleotide substitutions for each replicated SARS-CoV-1 genome (Eckerle et al. 2010). Here the process is annotated in two steps, nsp12- mediated misincorporation of a base (this reaction) and nsp14-mediated detection and removal of that base (next reaction).
Reaction - small molecule participants:
PPi [cytosol]
NTP(4-) [cytosol]
Reactome.org reaction link: R-HSA-9682563
======
Reaction input - small molecules:
nucleoside 5'-triphoshate(4-)
Reaction output - small molecules:
diphosphate(3-)
Reactome.org link: R-HSA-9682563