Reaction: nsp13 helicase melts secondary structures in SARS-CoV-2 genomic RNA template
- in pathway: Replication of the SARS-CoV-2 genome
nsp13 of SARS-CoV-2 possesses the nucleoside triphosphate hydrolase (NTPase) activity (Chen et al. 2020, Shu et al. 2020; Newman et al, 2021), and functions as an NTP-dependent RNA helicase that can unwind RNA helices (Shu et al. 2020; Mickolajczyk et al, 2021). Several G-quadruplex structures were confirmed in SARS-CoV-2 RNA and found to directly interact with nsp13, which may act to melt these structures (Ji et al. 2021).
nsp13 of SARS-CoV-1 is an ATP-dependent helicase that functions in the 5'-3' direction to unwind double stranded RNAs that have a 5' single strand overhang at least 20 nucleotides long. nsp13 can also act on double strand DNA in vitro, but dsRNA is thought to be its physiological substrate. The catalytic activity of SARS-CoV-1 nsp13 is increased in the presence of nsp12, the viral RNA-dependent RNA polymerase. nsp13 is needed for the replication of SARS-CoV-1 and is thought to act by melting secondary structures in the genomic RNA template during replication, and also to be involved in unwinding of RNA duplexes during transcription of viral genes. nsp13 is a promising target for experimental anti-SARS-CoV-1 drugs (Tanner et al. 2003, Ivanov et al. 2004, Bernini et al. 2006, Chen et al. 2009, Lee et al. 2010, Adedeji et al. 2012).
nsp13 of SARS-CoV-1 is an ATP-dependent helicase that functions in the 5'-3' direction to unwind double stranded RNAs that have a 5' single strand overhang at least 20 nucleotides long. nsp13 can also act on double strand DNA in vitro, but dsRNA is thought to be its physiological substrate. The catalytic activity of SARS-CoV-1 nsp13 is increased in the presence of nsp12, the viral RNA-dependent RNA polymerase. nsp13 is needed for the replication of SARS-CoV-1 and is thought to act by melting secondary structures in the genomic RNA template during replication, and also to be involved in unwinding of RNA duplexes during transcription of viral genes. nsp13 is a promising target for experimental anti-SARS-CoV-1 drugs (Tanner et al. 2003, Ivanov et al. 2004, Bernini et al. 2006, Chen et al. 2009, Lee et al. 2010, Adedeji et al. 2012).
Reaction - small molecule participants:
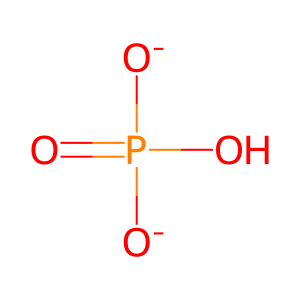
Pi [cytosol]
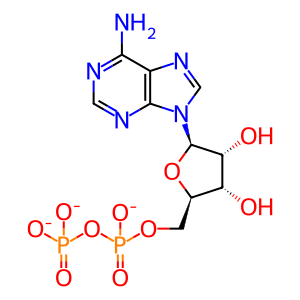
ADP [cytosol]
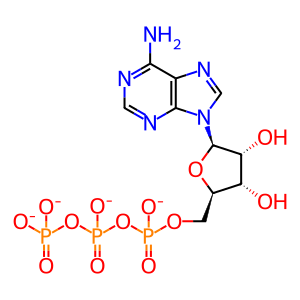
ATP [cytosol]
Reactome.org reaction link: R-HSA-9694265
======
Reaction input - small molecules:
ATP(4-)
Reaction output - small molecules:
hydrogenphosphate
ADP(3-)
Reactome.org link: R-HSA-9694265