Reaction: nsp14 acts as a 3'-to-5' exonuclease to remove misincorporated nucleotides from nascent RNA
- in pathway: Replication of the SARS-CoV-2 genome
In human SARS coronavirus 2 (SARS-CoV-2) nsp14 is essential for SARS-Cov-2 viability (Ogando et al, 2020). Mutations in nsp14 lead to an increased mutation load in the replicated viral genome, ultimately increasing genomic diversity (Eskier et al, 2020; Takada et al, 2020). The complex of nsp14 with nsp10 has 3'-5' exonuclease activity that is applied to mismatched nucleotides in newly replicated viral RNA (Saramago et al, 2021; Lin et al, 2021; Liu et al, 2021; Moeller et al, 2021; Ma et al, 2021). Binding to nsp10 increases the exonuclease activity of nsp14 260-fold (Riccio et al, 2022).
In SARS-CoV-1 nsp14 acts as 3'-5' exonuclease (Minskaia et al. 2006, Chen et al. 2007) that preferentially excises mismatched nucleotides from double stranded RNA (Minskaia et al. 2006, Bouvet et al. 2012). Binding to nsp10 increases the exonuclease activity of nsp14 (Bouvet et al. 2012, Subissi et al. 2014, Bouvet et al. 2014). nsp14 increases the fidelity of SARS-CoV-1 replication by the nsp12 RNA-dependent RNA polymerase by 21-fold (Eckerle et al. 2010).
In SARS-CoV-1 nsp14 acts as 3'-5' exonuclease (Minskaia et al. 2006, Chen et al. 2007) that preferentially excises mismatched nucleotides from double stranded RNA (Minskaia et al. 2006, Bouvet et al. 2012). Binding to nsp10 increases the exonuclease activity of nsp14 (Bouvet et al. 2012, Subissi et al. 2014, Bouvet et al. 2014). nsp14 increases the fidelity of SARS-CoV-1 replication by the nsp12 RNA-dependent RNA polymerase by 21-fold (Eckerle et al. 2010).
Reaction - small molecule participants:
NMP [cytosol]
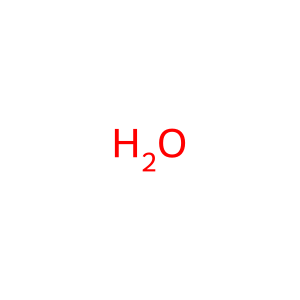
H2O [cytosol]
Reactome.org reaction link: R-HSA-9694632
======
Reaction input - small molecules:
water
Reaction output - small molecules:
ribonucleoside monophosphate
Reactome.org link: R-HSA-9694632