Reaction: nsp15 cleaves viral poly(U)-RNA
- in pathway: SARS-CoV-1 activates/modulates innate immune responses
Nonstructural protein 15 (nsp15) of severe acute respiratory syndrome coronavirus type 1 (SARS-CoV-1) is a nidoviral RNA uridylate‐specific endoribonuclease (EndoU) with the C‐terminal catalytic NendoU domain (Bhardwaj K et al. 2006; Ricagno S et al. 2006; Kim Y et al. 2020; reviewed in Deng X & Baker SC 2018). In vitro, the EndoU activity of nsp15 cleaves single-stranded (ss) and double-stranded (ds) RNAs with a preference for the 3′-ends of uridylates (Ivanov KA et al. 2004; Bhardwaj K et al. 2004, 2006). Nsp15 is thought to cleave RNA through a ribonuclease A (RNase A)-like mechanism producing 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl termini on the RNA products (Ricagno S et al. 2006; Bhardwaj K et al. 2006). The presence of Mn2+ enhanced nsp15 activity in vitro (Ivanov KA et al. 2004; Bhardwaj K et al. 2004). Biochemical studies revealed that SARS-CoV-1 nsp15 oligomerizes to form hexamers. The hexameric nsp15 binds RNA in solution (Guarino LA et al. 2005; Bhardwaj K et al. 2006). The crystal structures of nsp15 proteins from SARS-CoV-1, SARS-CoV-2, Middle East respiratory syndrome coronavirus (MERS-CoV) and murine hepatitis virus (MHV) confirmed that the biological unit of nsp15 is a hexamer (Ricagno S et al. 2006; Bhardwaj K et al. 2008; Xu X et la. 2006; Kim Y et al. 2020). The N-terminal domain (NTD) is important for oligomerization (Guarino LA et al. 2005; Joseph JS et al. 2007), and mutations that altered residues involved in subunit interactions generally resulted in the formation of catalytically inactive monomers (Bhardwaj K et al. 2008). MHV nsp15 has been shown to co-localize and interact with the replication/transcription complex (RTC)-associated proteins nsp8 and nsp12 in murine 17Cl-1 cells infected with MHV (Athmer J et al. 2017). Further, MERS-CoV nsp15 interacted with Nsp8 during both the pulldown and MST assays suggesting that coronavirus nsp15 functions in conjunction with the viral RTC (Zhang L et al. 2018). Though nsp15 is associated with the viral transcription/replication machinery, the role of nsp15 in viral replication remains unclear. MHV or human coronavirus 229E (HCoV-229E) that contain a catalytically-deficient nsp15 mutants stimulated MDA5 (IFIH1)-mediated IFN production and activated host dsRNA sensors such as PKR and RNase L in human and mouse cells (Kindler E et al. 2017; Deng X et al. 2017). The data suggest that SARS-CoV-1 nsp15 binds and cleaves viral polyuridine (polyU) RNA sequences degrading dsRNA at the site of viral RNA synthesis. The nsp15-mediated cleavage of RNA is thought to prevent an activation of host dsRNA sensors.
This Reactome event shows SARS-CoV-1 nsp15-mediated cleavage of viral poly(U) dsRNA and ssRNA substrates within the viral RTC.
Reaction - small molecule participants:
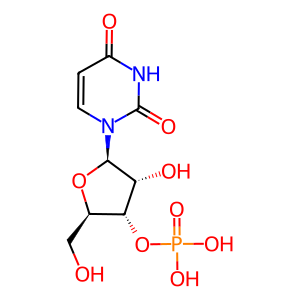
3'-UMP [cytosol]
Reactome.org reaction link: R-HSA-9705961
======
Reaction input - small molecules:
Reaction output - small molecules:
3'-UMP
Reactome.org link: R-HSA-9705961