Reaction: nsp15 cleaves viral poly(U)-RNA
- in pathway: Replication of the SARS-CoV-2 genome
Nonstructural protein 15 of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2 nsp15) is a nidoviral RNA uridylate‐specific endoribonuclease (EndoU) with the C‐terminal catalytic NendoU domain (Kim Y et al. 2020, 2021; Frazier MN et al. 2021). SARS-CoV-2 nsp15 shares 88% sequence identity and 95% sequence similarity with nsp15 of SARS-CoV-1 (Kim Y et al. 2020). Similar to its SARS-CoV-1 orthologue, nsp15 of SARS-CoV-2 forms a hexamer that consists of a dimer of trimers (Kim Y et al. 2020; Pillon MC et al. 2021). Nsp15 proteins from Middle East respiratory syndrome coronavirus (MERS-CoV) and murine hepatitis virus (MHV) also function as hexamers (Ricagno S et al. 2006; Bhardwaj K et al. 2008; Xu X et la. 2006; Kim Y et al. 2020). In vitro, the EndoU activity of SARS-CoV-1 nsp15 cleaves single-stranded (ss) and double-stranded (ds) RNAs with a preference for the 3′-ends of uridylates (Ivanov KA et al. 2004; Bhardwaj K et al. 2004, 2006). SARS-CoV-2 nsp15 was found to bind and hydrolyze 4, 7, and 20 nucleotide long RNA (Kim Y et al. 2021). Structural alignment of the catalytic site residues and RNA ligands of SARS-CoV-2 nsp15 and ribonuclease A (RNase A) suggests that nsp15 cleaves RNA through a ribonuclease A (RNase A)-like mechanism producing 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl termini on the RNA products (Kim Y et al. 2021). Mutations in the SARS-CoV-2 nsp15 catalytic site resulted in reduced or abrogated RNA cleavage (Frazier MN et al. 2021). The presence of Mn2+ enhanced nsp15 activity in vitro (Kim Y et al. 2021). Biochemical studies with nsp15 originated from MERS-CoV or MHV suggest that coronavirus nsp15 functions in conjunction with the viral RTC (Zhang L et al. 2018; Athmer J et al. 2017). Further, MHV or human coronavirus 229E (HCoV-229E) that contain a catalytically-deficient nsp15 mutants stimulated MDA5 (IFIH1)-mediated IFN production and activated host dsRNA sensors such as PKR and RNase L in human and mouse cells (Kindler E et al. 2017; Deng X et al. 2017). Together these data suggest that coronavirus nsp15 proteins, including SARS-CoV-2 nsp15, bind and cleave viral polyuridine (polyU) RNA sequences degrading dsRNA at the site of viral RNA synthesis. The nsp15-mediated cleavage of RNA is thought to prevent an activation of host dsRNA sensors dampening IFN production in mammalian cells (Kindler E et al. 2017; Deng X et al. 2017; Hackbar M et al. 2020).
This Reactome event shows SARS-CoV-2 nsp15-mediated cleavage of viral poly(U) dsRNA and ssRNA substrates within the viral RTC.
Reaction - small molecule participants:
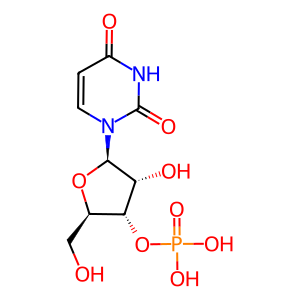
3'-UMP [cytosol]
Reactome.org reaction link: R-HSA-9755252
======
Reaction input - small molecules:
Reaction output - small molecules:
3'-UMP
Reactome.org link: R-HSA-9755252