Reaction: Formation of the Spliceosomal C* complex
- in pathway: mRNA Splicing - Major Pathway
DHX38 (homolog of yeast Prp16) hydrolyzes ATP to remodel the Spliceosomal C complex to the Spliceosomal C* complex (Ortlepp et al. 1998, Zhou and Reed 1998), which will ligate the exons and produce the intron lariat, yielding the Spliceosomal P (post-catalytic) complex. DHX38 appears to dissociate shortly after remodeling the Spliceosomal C complex (Zhang et al. 2017, Kastner et al. 2019). Catalysis of exon ligation will occur in the same active site as branching occurred. Therefore, the pre-mRNA must be moved relative to the active site. DHX38 binds the pre-mRNA downstream of the intron branch point, displaces YJU2 and CWC25, and rotates the branch helix out of the active site to allow the 3' splice junction to enter (Bertram et al. 2017, Zhang et al. 2017). The branch helix is stabilized after re-positioning by CDC40 (homolog of yeast Prp17) bound to the 5' stem-loop of the U6 snRNA and PRPF8 (homolog of yeast Prp8) of the U5 snRNP (Bertram et al. 2017, Zhang et al. 2017). SLU7 and PRPF18 (homolog of yeast Prp18) are components of the Spliceosomal C* complex and participate in docking the 3' splice site in the active site (Bertram et al. 2017, Zhang et al. 2017). The 3' end of the upstream exon base-pairs with loop I of U5 snRNA and the branch point region of the intron base-pairs with the U2 snRNA and U6 snRNA (Bertram et al. 2017, Zhang et al. 2017). As in yeast (Wilkinson et al. 2017), the 3' splice site may form non-Watson-Crick base pairs with the 5' splice site and branch point of the intron. The 3' splice site may also interact with SLU7 (Bertram et al. 2017, Zhang et al. 2017), PRPF18 (homolog of yeast Prp18), and DHX8 (homolog of yeast Prp22). As in the Bact complex, the catalytic magnesium ions of the active Spliceosomal C complex are coordinated by phosphates of the U6 snRNA (Bertram et al. 2017, Zhang et al. 2017). The components of the Spliceosomal C* complex have been ascertained using proteomic and structural methods (Jurica et al. 2002, Makarov et al. 2002, Rappsilber et al. 2002, Ilagan et al. 2013, Bertram et al. 2017, Zhang et al. 2017, Kastner et al. 2019, also inferred from yeast homologs in Fica et al. 2017). Mutations in DHX38 affect splicing and can cause retinitis pigmentosa (Ajmal et al. 2014, Obuca et al. 2022).
Reaction - small molecule participants:
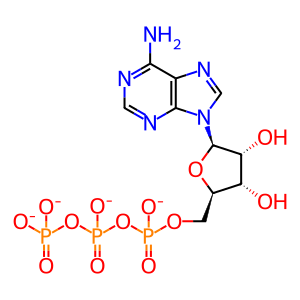
ATP [nucleoplasm]
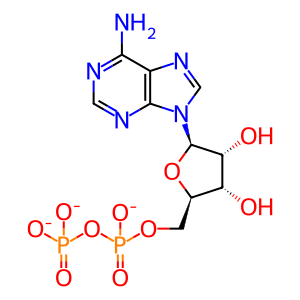
ADP [nucleoplasm]
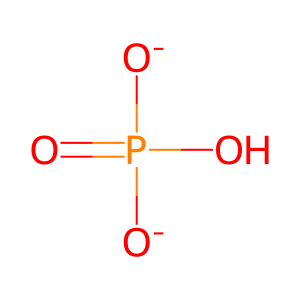
Pi [nucleoplasm]
Reactome.org reaction link: R-HSA-9770141
======
Reaction input - small molecules:
ATP(4-)
Reaction output - small molecules:
ADP(3-)
hydrogenphosphate
Reactome.org link: R-HSA-9770141