Reaction: DNMT1,3A,3B:PRC2 methylates cytosine and histone H3
- in pathway: PRC2 methylates histones and DNA
DNA methyltransferases (DNMTs) associate with EZH2 of the Polycomb Repressive Complex 2 (PRC2) and methylate the 5 position of the cytosine ring in DNA (Vire et al. 2006). The histone methyltransferase activity of EZH2 also trimethylates lysine-27 of histone H3 (H3K27me3). The promoters of the MYT, WNT1, KCNA1, and CNR1 genes are methylated on cytosine by the DNMT:PRC2 complex however not all loci that have H3K27me3 by PRC2 also have cytosine methylation (Vire et al. 2006, Brinkman et al. 2012). DNA methylation and H3K27me3 appear to be mutually exclusive in CpG islands but are compatible throughout most of the rest of the genome (Brinkman et al. 2012). In mouse, DNA methylation and H3K27me3 appear to be antagonistic at most loci: loss of DNA methylation causes increased H3K27me3 while loss of PRC2 has little effect on DNA methylation (Hagarman et al. 2013). By competing with DNMT3a,b for association with PRC2, DNMT3L may antagonize DNA methylation at sites bound by PRC2 (Neri et al. 2013).
Reaction - small molecule participants:
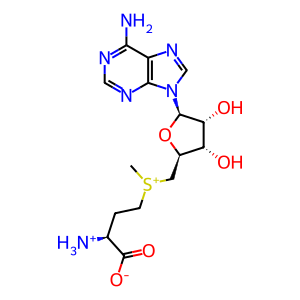
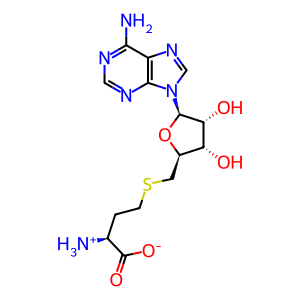
AdoHcy [nucleoplasm]
AdoMet [nucleoplasm]
Reactome.org reaction link: R-HSA-212269
======
Reaction input - small molecules:
S-adenosyl-L-methionine zwitterion
Reaction output - small molecules:
S-adenosyl-L-homocysteine zwitterion
Reactome.org link: R-HSA-212269