Reaction: WHSC1 (KMT3G), NSD1 (KMT3B), SMYD2 (KMT3C) methylate lysine-37 of histone H3 (H3K36)
- in pathway: PKMTs methylate histone lysines
Methylation of histone H3 lysine-37 (H3K36) is tightly associated with actively transcribed genes and appears to correspond primarily with coding regions (Wagner & Carpenter 2011).
WHSC1 (KMT3G, NSD2, MMSET), a member of the SET2 family, dimethylates H3K36 when provided with nucleosome substrates (Li et al 2009; Qiao et al. 2011). Dimethylation of histone H3 at lysine-37 (H3K36me2) is thought to be the principal chromatin-regulatory activity of WHSC1 (Kuo et al. 2011), SMYD2 (KMT3C) (Brown et al 2006) and NSD1 (KMT3B) (Li et al. 2009, Qiao et al. 2011).
WHSC1 (KMT3G, NSD2, MMSET), a member of the SET2 family, dimethylates H3K36 when provided with nucleosome substrates (Li et al 2009; Qiao et al. 2011). Dimethylation of histone H3 at lysine-37 (H3K36me2) is thought to be the principal chromatin-regulatory activity of WHSC1 (Kuo et al. 2011), SMYD2 (KMT3C) (Brown et al 2006) and NSD1 (KMT3B) (Li et al. 2009, Qiao et al. 2011).
Reaction - small molecule participants:
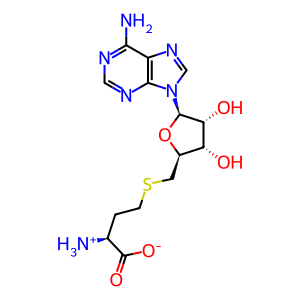
AdoHcy [nucleoplasm]
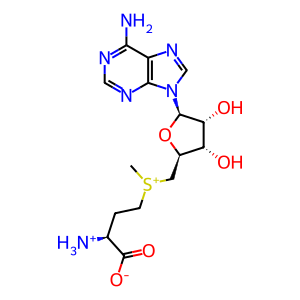
AdoMet [nucleoplasm]
Reactome.org reaction link: R-HSA-4827383
======
Reaction input - small molecules:
S-adenosyl-L-methionine zwitterion
Reaction output - small molecules:
S-adenosyl-L-homocysteine zwitterion
Reactome.org link: R-HSA-4827383