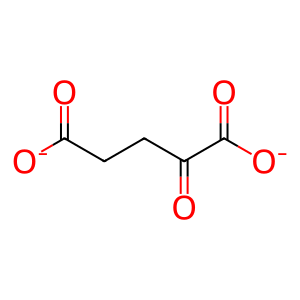
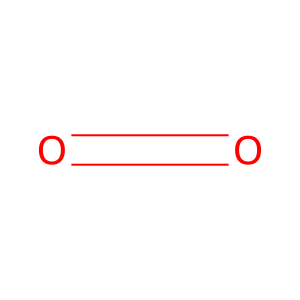
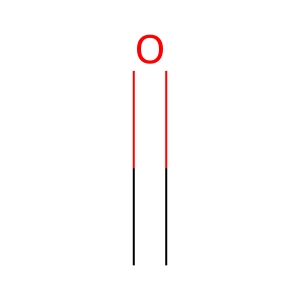
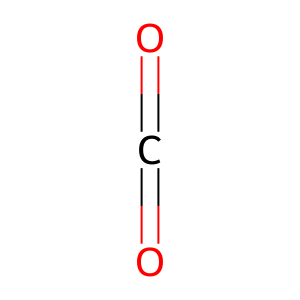
Reaction: KDM5A-D demethylate Me3K5-histone H3
- in pathway: HDMs demethylate histones
All characterized lysine demethylases other than KDM1A belong to the jumonjiC domain (JmjC) containing family.The JmjC KDMs are members of the Cupin superfamily of mononuclear Fe (II) dependent oxygenases, which are characterized by the presence of a double-stranded beta-helix core fold. The JmjC KDMs require 2 oxoglutarate (2 OG) and molecular oxygen as co substrates, producing, along with formaldehyde, succinate and carbon dioxide. This hydroxylation based mechanism does not require a protonatable lysine epsilon-amine group and consequently JmjC containing demethylases are able to demethylate tri , di and monomethylated lysines.
KDM5A-D (JARID1A-D) catalyse the demethylation of di- or tri-methylated lysine-5 of histone H3 (H3K4Me2/3) (Christensen et al. 2007, Klose et al. 2007, Lee et al. 2007, Secombe et al. 2007, Seward et al. 2007, Iwase et al. 2007).
Reaction - small molecule participants:
CH2O [nucleoplasm]
CO2 [nucleoplasm]
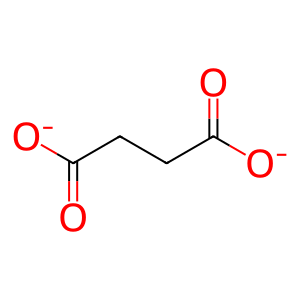
SUCCA [nucleoplasm]
2OG [nucleoplasm]
O2 [nucleoplasm]
Reactome.org reaction link: R-HSA-5661116
======
Reaction input - small molecules:
2-oxoglutarate(2-)
dioxygen
Reaction output - small molecules:
formaldehyde
carbon dioxide
succinate(2-)
Reactome.org link: R-HSA-5661116