Reaction: RHOA GEFs activate RHOA
- in pathway: RHOA GTPase cycle
The following guanine nucleotide exchange factors (GEFs) were shown to bind RHOA and catalyze GDP to GTP exchange on RHOA, resulting in formation of the active RHOA:GTP complex (the high throughput study by Bagci et al. 2020 looked at preferential binding of GEFs to nucleotide-free RHOA mutant compared to wild type RHOA but did not examine guanine nucleotide exchange):
AKAP13 (Zheng et al. 1995; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF1 (Hart et al. 1996; Jaiswal et al. 2011; Jaiswal et al. 2013; Toffali et al. 2017; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF2 (Krendel et al. 2002; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF3 (Arthur et al. 2002; Sloan et al. 2012; Müller et al. 2020)
ARHGEF5 (Xie et al. 2005; Wang et al. 2009; supported by Bagci et al. 2020)
ARHGEF10 (Mohl et al. 2006; Aoki et al. 2009; Müller et al. 2020)
ARHGEF11 (Rümenapp et al. 1999; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF12 (Reuther et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF15 (Fukushima et al. 2016; Müller et al. 2020)
ARHGEF17 (Rümenapp et al. 2002; Mitin et al. 2012; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF19 (Wang et al. 2004; Müller et al. 2020)
ARHGEF25 (Sloan et al. 2012; Müller et al. 2020)
ARHGEF28 (van Horck et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020)
ECT2 (Tatsumoto et al. 1999; Müller et al. 2020; supported by Bagci et al. 2020)
MCF2 (Sloan et al. 2012; Jaiswal et al. 2013; Müller et al. 2020)
NET1 (Alberts and Treisman 1998; Müller et al. 2020)
NGEF (Zhang et al. 2007; Müller et al. 2020; supported by Bagci et al. 2020)
OBSCN (Ford Speelman et al. 2009)
PLEKHG5 (de Toledo et al. 2001; Liu and Horowitz 2006; Müller et al. 2020)
PLEKHG6 (Müller et al. 2020)
RASGRF2 (Müller et al. 2020)
TRIO (TRIO and KALRN possess two GEF domains, the N-terminal GEF1 and the C-terminal GEF2; Debant et al. 1996 demonstrated activation of RHOA by the C-terminal GEF2 domain of TRIO, while Peurois et al. 2017 demonstrated activation of RHOA by the N-terminal GEF1 domain of TRIO in the presence of membranes, Williams et al. 2007 showed that in C-elegans GNAQ-mediated activation of TRIO results in RHOA activation that drived locomotion and egg laying, and Bagci et al. 2020 reported binding of inactive RHOA mutant to the full-length TRIO; Debant et al. 1996 and Jaiswal et al. 2013 showed that the N-terminal GEF1 domain of TRIO does not activate RHOA; Müller et al. 2020 detected no activation of RHOA in the presence of full-length TRIO, but in this study HEK293 cells were used, which were previously shown by Williams et al. 2007 to have a low TRIO-activation ability downstream of GNAQ)
VAV1 (Heo et al. 2005; Müller et al. 2020)
VAV2 (Heo et al. 2005; Sloan et al. 2012; Schuebel et al. 1998; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
VAV3 (Movilla and Bustelo 1999, Sachdev et al. 2002; Müller et al. 2020)
They are annotated as RHOA GEF set members.
The following GEFs were shown to bind and activate RHOA in some but not all studies, or have not been sufficiently studied, and are annotated as candidate RHOA GEFs:
ABR (Chuang et al. 1995, Manser et al. 1998: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF4 (Müller et al. 2020: activation of RHOA; Gotthardt and Ahmadian 2007, Anderson and Hamann 2012, Jaiswal et al. 2013: no activation of RHOA)
ARHGEF7 (Manser et al. 1998: activation of RHOA; Jones and Katan 2007, Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
ARHGEF10L (Winkler et al. 2005: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF18 (Niu et al. 2003, Blomquist et al. 2000: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF40 (Curtis et al. 2004: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
BCR (Chuang et al. 1995: activation of RHOA; Bagci et al. 2020: binding to RHOA; Müller et al. 2020: no activation of RHOA)
DEF6 (Mavrakis et al. 2004: probable GEF, in vitro GEF activity not tested)
DOCK2 (Toffali et al. 2017: inconclusive; Müller et al. 2020: no activation of RHOA)
FARP1 (Koyano et al. 2001: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: inconclusive binding to RHOA)
KALRN (Penzes et al. 2001: the C-terminal GEF2 domain of KALRN binds RHOA and activates it, although activation was not demonstrated in vitro, while the N-terminal GEF1 domain of KALRN does not bind to RHOA; Wu et al. 2013: no effect on RHOA activity in mice with heterozygous Kalrn gene knockout; Müller et al. 2020: no activation of RHOA by full-length KALRN)
MCF2L (Whitehead et al. 1999, Sloan et al. 2012, Jaiswal et al. 2013, Müller et al. 2020: activation of RHOA; Bagci et al. 2020: no binding to RHOA)
PLEKHG3 (Bagci et al. 2020: binding to RHOA, RHOA activation not tested; Müller et al. 2020: no activation of RHOA)
PLEKHG4 (Gupta et al. 2013: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
PREX1 (Jaiswal et al. 2013: activation of RHOA; Marei et al. 2016, Müller et al. 2020: no activation of RHOA)
PREX2 (Müller et al. 2020: activation of RHOA; Joseph and Norris 2005: no activation of RHOA)
TIAM1 (Michiels et al. 1995: activation of RHOA; Jaiswal et al. 2013, Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
The following GEFs do not act on RHOA or have been reported to not bind to RHOA, as indicated, without testing for activity:
ALS2 (Müller et al. 2020)
ARHGEF6 (Ramakers et al. 2012; Müller et al. 2020)
ARHGEF9 (Reid et al. 1999; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF16 (Hiramoto Yamaki et al. 2010; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ARHGEF26 (Ellerbroek et al. 2004; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ARHGEF39 (Müller et al. 2020)
DNMBP (Jaiswal et al. 2013; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK1 (Côté and Vuori 2002; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK3 (Müller et al. 2020)
DOCK4 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK5 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK6 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK7 (Müller et al. 2020; no binding to RHOA byBagci et al. 2020)
DOCK8 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK9 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK10 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK11 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ECT2L (Müller et al. 2020)
FARP2 (Kubo et al. 2002; Müller et al. 2020);
FGD1 (Olson et al. 1996; Müller et al. 2020)
FGD2 (Huber et al. 2008; Müller et al. 2020)
FGD3 (Müller et al. 2020)
FGD5 (Müller et al. 2020)
FGD6 (Müller et al. 2020)
FGD4 (Umikawa et al. 1999; Müller et al. 2020)
GNA13 (Teo et al. 2016)
ITSN1 (Hussain et al. 2001; Jaiswal et al. 2013; Müller et al. 2020)
ITSN2 (Müller et al. 2020)
MCF2L2 (Müller et al. 2020)
PLEKHG1 (Abiko et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
PLEKHG2 (Ueda et al. 2008; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
PLEKHG4B (Müller et al. 2020)
SOS1 (Nimnual et al. 1998; Müller et al. 2020)
SOS2 (Nimnual et al. 1998; Müller et al. 2020)
SPATA13 (Hamann et al. 2007; Kawasaki et al. 2007; Bristow et al. 2009; Müller et al. 2020)
SWAP70 (Ocana Morgner et al. 2009; no binding to RHOA by Bagci et al. 2020)
TIAM2 (Müller et al. 2020)
AKAP13 (Zheng et al. 1995; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF1 (Hart et al. 1996; Jaiswal et al. 2011; Jaiswal et al. 2013; Toffali et al. 2017; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF2 (Krendel et al. 2002; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF3 (Arthur et al. 2002; Sloan et al. 2012; Müller et al. 2020)
ARHGEF5 (Xie et al. 2005; Wang et al. 2009; supported by Bagci et al. 2020)
ARHGEF10 (Mohl et al. 2006; Aoki et al. 2009; Müller et al. 2020)
ARHGEF11 (Rümenapp et al. 1999; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF12 (Reuther et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF15 (Fukushima et al. 2016; Müller et al. 2020)
ARHGEF17 (Rümenapp et al. 2002; Mitin et al. 2012; Müller et al. 2020; supported by Bagci et al. 2020)
ARHGEF19 (Wang et al. 2004; Müller et al. 2020)
ARHGEF25 (Sloan et al. 2012; Müller et al. 2020)
ARHGEF28 (van Horck et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020)
ECT2 (Tatsumoto et al. 1999; Müller et al. 2020; supported by Bagci et al. 2020)
MCF2 (Sloan et al. 2012; Jaiswal et al. 2013; Müller et al. 2020)
NET1 (Alberts and Treisman 1998; Müller et al. 2020)
NGEF (Zhang et al. 2007; Müller et al. 2020; supported by Bagci et al. 2020)
OBSCN (Ford Speelman et al. 2009)
PLEKHG5 (de Toledo et al. 2001; Liu and Horowitz 2006; Müller et al. 2020)
PLEKHG6 (Müller et al. 2020)
RASGRF2 (Müller et al. 2020)
TRIO (TRIO and KALRN possess two GEF domains, the N-terminal GEF1 and the C-terminal GEF2; Debant et al. 1996 demonstrated activation of RHOA by the C-terminal GEF2 domain of TRIO, while Peurois et al. 2017 demonstrated activation of RHOA by the N-terminal GEF1 domain of TRIO in the presence of membranes, Williams et al. 2007 showed that in C-elegans GNAQ-mediated activation of TRIO results in RHOA activation that drived locomotion and egg laying, and Bagci et al. 2020 reported binding of inactive RHOA mutant to the full-length TRIO; Debant et al. 1996 and Jaiswal et al. 2013 showed that the N-terminal GEF1 domain of TRIO does not activate RHOA; Müller et al. 2020 detected no activation of RHOA in the presence of full-length TRIO, but in this study HEK293 cells were used, which were previously shown by Williams et al. 2007 to have a low TRIO-activation ability downstream of GNAQ)
VAV1 (Heo et al. 2005; Müller et al. 2020)
VAV2 (Heo et al. 2005; Sloan et al. 2012; Schuebel et al. 1998; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
VAV3 (Movilla and Bustelo 1999, Sachdev et al. 2002; Müller et al. 2020)
They are annotated as RHOA GEF set members.
The following GEFs were shown to bind and activate RHOA in some but not all studies, or have not been sufficiently studied, and are annotated as candidate RHOA GEFs:
ABR (Chuang et al. 1995, Manser et al. 1998: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF4 (Müller et al. 2020: activation of RHOA; Gotthardt and Ahmadian 2007, Anderson and Hamann 2012, Jaiswal et al. 2013: no activation of RHOA)
ARHGEF7 (Manser et al. 1998: activation of RHOA; Jones and Katan 2007, Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
ARHGEF10L (Winkler et al. 2005: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF18 (Niu et al. 2003, Blomquist et al. 2000: activation of RHOA; Müller et al. 2020: no activation of RHOA)
ARHGEF40 (Curtis et al. 2004: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
BCR (Chuang et al. 1995: activation of RHOA; Bagci et al. 2020: binding to RHOA; Müller et al. 2020: no activation of RHOA)
DEF6 (Mavrakis et al. 2004: probable GEF, in vitro GEF activity not tested)
DOCK2 (Toffali et al. 2017: inconclusive; Müller et al. 2020: no activation of RHOA)
FARP1 (Koyano et al. 2001: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: inconclusive binding to RHOA)
KALRN (Penzes et al. 2001: the C-terminal GEF2 domain of KALRN binds RHOA and activates it, although activation was not demonstrated in vitro, while the N-terminal GEF1 domain of KALRN does not bind to RHOA; Wu et al. 2013: no effect on RHOA activity in mice with heterozygous Kalrn gene knockout; Müller et al. 2020: no activation of RHOA by full-length KALRN)
MCF2L (Whitehead et al. 1999, Sloan et al. 2012, Jaiswal et al. 2013, Müller et al. 2020: activation of RHOA; Bagci et al. 2020: no binding to RHOA)
PLEKHG3 (Bagci et al. 2020: binding to RHOA, RHOA activation not tested; Müller et al. 2020: no activation of RHOA)
PLEKHG4 (Gupta et al. 2013: activation of RHOA; Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
PREX1 (Jaiswal et al. 2013: activation of RHOA; Marei et al. 2016, Müller et al. 2020: no activation of RHOA)
PREX2 (Müller et al. 2020: activation of RHOA; Joseph and Norris 2005: no activation of RHOA)
TIAM1 (Michiels et al. 1995: activation of RHOA; Jaiswal et al. 2013, Müller et al. 2020: no activation of RHOA; Bagci et al. 2020: no binding to RHOA)
The following GEFs do not act on RHOA or have been reported to not bind to RHOA, as indicated, without testing for activity:
ALS2 (Müller et al. 2020)
ARHGEF6 (Ramakers et al. 2012; Müller et al. 2020)
ARHGEF9 (Reid et al. 1999; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF16 (Hiramoto Yamaki et al. 2010; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ARHGEF26 (Ellerbroek et al. 2004; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ARHGEF39 (Müller et al. 2020)
DNMBP (Jaiswal et al. 2013; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK1 (Côté and Vuori 2002; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK3 (Müller et al. 2020)
DOCK4 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK5 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK6 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK7 (Müller et al. 2020; no binding to RHOA byBagci et al. 2020)
DOCK8 (Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK9 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK10 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
DOCK11 (Ruiz Lafuente et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
ECT2L (Müller et al. 2020)
FARP2 (Kubo et al. 2002; Müller et al. 2020);
FGD1 (Olson et al. 1996; Müller et al. 2020)
FGD2 (Huber et al. 2008; Müller et al. 2020)
FGD3 (Müller et al. 2020)
FGD5 (Müller et al. 2020)
FGD6 (Müller et al. 2020)
FGD4 (Umikawa et al. 1999; Müller et al. 2020)
GNA13 (Teo et al. 2016)
ITSN1 (Hussain et al. 2001; Jaiswal et al. 2013; Müller et al. 2020)
ITSN2 (Müller et al. 2020)
MCF2L2 (Müller et al. 2020)
PLEKHG1 (Abiko et al. 2015; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
PLEKHG2 (Ueda et al. 2008; Müller et al. 2020; no binding to RHOA by Bagci et al. 2020)
PLEKHG4B (Müller et al. 2020)
SOS1 (Nimnual et al. 1998; Müller et al. 2020)
SOS2 (Nimnual et al. 1998; Müller et al. 2020)
SPATA13 (Hamann et al. 2007; Kawasaki et al. 2007; Bristow et al. 2009; Müller et al. 2020)
SWAP70 (Ocana Morgner et al. 2009; no binding to RHOA by Bagci et al. 2020)
TIAM2 (Müller et al. 2020)
Reaction - small molecule participants:
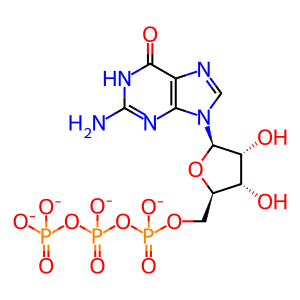
GTP [cytosol]
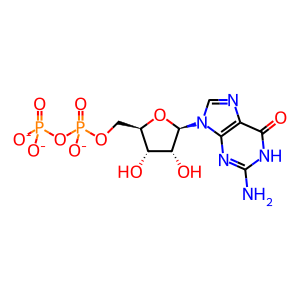
GDP [cytosol]
Reactome.org reaction link: R-HSA-8980691
======
Reaction input - small molecules:
GTP(4-)
Reaction output - small molecules:
GDP(3-)
Reactome.org link: R-HSA-8980691