Reaction: CDC42 GEFs activate CDC42
- in pathway: CDC42 GTPase cycle
The following guanine nucleotide exchange factors (GEFs) were shown to bind CDC42 and catalyze GDP to GTP exchange on CDC42, resulting in formation of the active CDC42:GTP complex (the high throughput study by Bagci et al. 2020 examined binding of GEFs to inactive CDC42 mutants without testing for CDC42-directed GEF activity and is cited as supporting evidence):
ARHGEF4 (also known as Asef) (Itoh et al. 2008; Anderson and Hamann 2012; Gotthardt and Ahmadian 2007; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF9 (also known as hPEM2) (Reid et al. 1999; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF15 (Fukushima et al. 2016; Müller et al. 2020)
DEF6 (Mavrakis et al. 2004)
DNMBP (also known as Tuba) (Salazar et al. 2003; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
FGD1 (Olson et al. 1996; Müller et al. 2020)
FGD2 (Huber et al. 2008; Müller et al. 2020)
FGD3 (Hayakawa et al. 2008; Müller et al. 2020)
FGD4 (Umikawa et al. 1999; Müller et al. 2020)
MCF2 (Ueda et al. 2004; Jaiswal et al. 2013; Müller et al. 2020)
MCF2L (Ueda et al. 2004; Whitehead et al. 1999; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG1 (Abiko et al. 2015; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG3 (Nguyen et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG4 (Gupta et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG4B (Müller et al. 2020)
SPATA13 (Hamann et al. 2007; Kawasaki et al. 2007; Bristow et al. 2009; Müller et al. 2020)
The following GEFs; annotated as CDC42 candidate GEFs; were shown to activate CDC42 by some but not all studies (the high throughput study by Bagci et al. 2020 examined binding of GEFs to inactive CDC42 mutants without testing for CDC42-directed GEF activity and is cited as supporting evidence):
ABR (Chuang et al. 1995: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF5 (Xie et al. 2005: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF6 (also known as alphaPIX or KIAA006) (Manser et al. 1998, Ramakers et al. 2012, Meseke et al. 2013: CDC42-directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF10 (Müller et al. 2020: CDC42 directed GEF activity; Mohl et al. 2006: no CDC42-directed GEF activity)
ARHGEF11 (Bagci et al. 2020: binding to inactive CDC42; Rümenapp et al. 1999, Jaiswal et al. 2011, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF12 (Bagci et al. 2020: binding to inactive CDC42; Reuther et al. 2001, Jaiswal et al. 2011, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF16 (Bagci et al. 2020: binding to inactive CDC42; Hiramoto Yamaki et al. 2010, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF19 (Wang et al. 2004: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF25 (Guo et al. 2003: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF26 (Müller et al. 2020: CDC42 directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Ellerbroek et al. 2004: no CDC42 directed GEF activity)
BCR (Chuang et al. 1995; Korus et al. 2002: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK6 (Miyamoto et al. 2007: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK7 (Kukimoto Niino et al. 2019, Wilkes et al. 2014, Zhou et al. 2013, Yamauchi et al. 2008: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK8 (Ruusala and Aspenstrom 2004, Harada et al. 2012, Martins et al. 2016, Xu et al. 2017: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK9 (also known as Zizimin1) (Meller et al. 2002, Cote and Vuori 2002, Kulkarni et al. 2011: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK10 (Gadea et al. 2008, Ruiz Lafuente et al. 2015: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK11 (Lin et al. 2006: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
ECT2 (Tatsumoto et al. 1999, Fortin et al. 2012: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
FARP1 (Amado Azevedo et al. 2017: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
GNA13 (Yan et al. 2015: GNA13 increases the amount of GTP-bound CDC42, but the evidence for direct effect is lacking)
ITSN1 (Hussain et al. 2001, Jaiswal et al. 2013: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
NGEF (Zhang et al. 2007: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
PLEKHG2 (Ueda et al. 2008: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
PREX1 (Jaiswal et al. 2013: CDC42 directed GEF activity; Marei et al. 2016, Müller et al. 2020: no CDC42 directed GEF activity)
PREX2 (Müller et al. 2020: CDC42-directed GEF activity; Joseph and Norris 2005: no CDC42-directed GEF activity)
RASGRF2 (Müller et al. 2020: CDC42-directed GEF activity; Calvo et al. 2011: no CDC42-directed GEF activity)
TIAM1 (Michiels et al. 1995: CDC42 directed GEF activity; Itoh et al. 2008, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
TRIO (Peurois et al. 2017, Fortin et al. 2012, Jaiswal et al. 2013: CDC42 directed GEF activity, the presence of membrane may be necessary; Debant et al. 1996: no CDC42-directed GEF activity of either the N-terminal GEF1 domain or the C-terminal GEF2 domain of TRIO in vitro; Müller et al. 2020: no CDC42 directed GEF activity of the full-length TRIO; Bagci et al. 2020: no binding of full-length TRIO to inactive CDC42)
VAV2 (Itoh et al. 2008, Aoki et al. 2005, Heo et al. 2005, Jaiswal et al. 2013: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42 directed GEF activity)
VAV3 (Sachdev et al. 2002; Aoki et al. 2005: CDC42 directed GEF activity; Movilla and Bustelo 1999, Müller et al. 2020: no CDC42 directed GEF activity)
The following GEFs do not act on CDC42 or were shown to not bind to inactive CDC42 mutant in the high throughput screen by Bagci et al. 2020):
AKAP13 (Zheng et al. 1995; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ALS2 (Müller et al. 2020)
ARHGEF1 (Hart et al. 1996; Jaiswal et al. 2013; Jaiswal et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF2 (Krendel et al. 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF3 (Arthur et al. 2002; Müller et al. 2020)
ARHGEF7 (Manser et al. 1998; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF10L (Winkler et al. 2005; Müller et al. 2020)
ARHGEF17 (Rümenapp et al. 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF18 (Niu et al. 2003; Blomquist et al. 2000; Müller et al. 2020)
ARHGEF28 (van Horck et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF39 (Müller et al. 2020)
ARHGEF40 (Curtis et al. 2004; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK1 (Cote and Vuori 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK2 (Kulkarni et al. 2011; Kwofie and Skowronski 2008; Müller et al. 2020)
DOCK3 (Kwofie and Skowronski 2008; Müller et al. 2020)
DOCK4 (Kwofie and Skowronski 2008; Abraham et al. 2015; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK5 (Vives et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ECT2L (Müller et al. 2020)
FARP2 (Kubo et al. 2002; Müller et al. 2020)
FGD5 (Müller et al. 2020)
FGD6 (Müller et al. 2020)
ITSN2 (Müller et al. 2020)
KALRN (Penzes et al. 2001; Müller et al. 2020)
MCF2L2 (Müller et al. 2020)
NET1 (Alberts and Treisman 1998; Müller et al. 2020)
OBSCN (Ford Speelman et al. 2009)
PLEKHG5 (De Toledo et al. 2001; Müller et al. 2020)
PLEKHG6 (Müller et al. 2020)
PLEKHG7 (Müller et al. 2020)
RASGRF1 (Müller et al. 2020)
SOS1 (Nimnual et al. 1998; Müller et al. 2020)
SOS2 (Nimnual et al. 1998; Müller et al. 2020)
SWAP70 (Bagci et al. 2020: no binding to inactive CDC42)
TIAM2 (Müller et al. 2020)
VAV1 (Aghazadeh et al. 2000; Müller et al. 2020)
ARHGEF4 (also known as Asef) (Itoh et al. 2008; Anderson and Hamann 2012; Gotthardt and Ahmadian 2007; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF9 (also known as hPEM2) (Reid et al. 1999; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF15 (Fukushima et al. 2016; Müller et al. 2020)
DEF6 (Mavrakis et al. 2004)
DNMBP (also known as Tuba) (Salazar et al. 2003; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
FGD1 (Olson et al. 1996; Müller et al. 2020)
FGD2 (Huber et al. 2008; Müller et al. 2020)
FGD3 (Hayakawa et al. 2008; Müller et al. 2020)
FGD4 (Umikawa et al. 1999; Müller et al. 2020)
MCF2 (Ueda et al. 2004; Jaiswal et al. 2013; Müller et al. 2020)
MCF2L (Ueda et al. 2004; Whitehead et al. 1999; Jaiswal et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG1 (Abiko et al. 2015; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG3 (Nguyen et al. 2016; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG4 (Gupta et al. 2013; Müller et al. 2020; supported by Bagci et al. 2020)
PLEKHG4B (Müller et al. 2020)
SPATA13 (Hamann et al. 2007; Kawasaki et al. 2007; Bristow et al. 2009; Müller et al. 2020)
The following GEFs; annotated as CDC42 candidate GEFs; were shown to activate CDC42 by some but not all studies (the high throughput study by Bagci et al. 2020 examined binding of GEFs to inactive CDC42 mutants without testing for CDC42-directed GEF activity and is cited as supporting evidence):
ABR (Chuang et al. 1995: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF5 (Xie et al. 2005: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF6 (also known as alphaPIX or KIAA006) (Manser et al. 1998, Ramakers et al. 2012, Meseke et al. 2013: CDC42-directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF10 (Müller et al. 2020: CDC42 directed GEF activity; Mohl et al. 2006: no CDC42-directed GEF activity)
ARHGEF11 (Bagci et al. 2020: binding to inactive CDC42; Rümenapp et al. 1999, Jaiswal et al. 2011, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF12 (Bagci et al. 2020: binding to inactive CDC42; Reuther et al. 2001, Jaiswal et al. 2011, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF16 (Bagci et al. 2020: binding to inactive CDC42; Hiramoto Yamaki et al. 2010, Müller et al. 2020: no CDC42 directed GEF activity)
ARHGEF19 (Wang et al. 2004: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF25 (Guo et al. 2003: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
ARHGEF26 (Müller et al. 2020: CDC42 directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Ellerbroek et al. 2004: no CDC42 directed GEF activity)
BCR (Chuang et al. 1995; Korus et al. 2002: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK6 (Miyamoto et al. 2007: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK7 (Kukimoto Niino et al. 2019, Wilkes et al. 2014, Zhou et al. 2013, Yamauchi et al. 2008: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
DOCK8 (Ruusala and Aspenstrom 2004, Harada et al. 2012, Martins et al. 2016, Xu et al. 2017: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK9 (also known as Zizimin1) (Meller et al. 2002, Cote and Vuori 2002, Kulkarni et al. 2011: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK10 (Gadea et al. 2008, Ruiz Lafuente et al. 2015: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
DOCK11 (Lin et al. 2006: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
ECT2 (Tatsumoto et al. 1999, Fortin et al. 2012: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
FARP1 (Amado Azevedo et al. 2017: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
GNA13 (Yan et al. 2015: GNA13 increases the amount of GTP-bound CDC42, but the evidence for direct effect is lacking)
ITSN1 (Hussain et al. 2001, Jaiswal et al. 2013: CDC42-directed GEF activity; Müller et al. 2020: no CDC42-directed GEF activity)
NGEF (Zhang et al. 2007: CDC42 directed GEF activity; Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
PLEKHG2 (Ueda et al. 2008: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42-directed GEF activity)
PREX1 (Jaiswal et al. 2013: CDC42 directed GEF activity; Marei et al. 2016, Müller et al. 2020: no CDC42 directed GEF activity)
PREX2 (Müller et al. 2020: CDC42-directed GEF activity; Joseph and Norris 2005: no CDC42-directed GEF activity)
RASGRF2 (Müller et al. 2020: CDC42-directed GEF activity; Calvo et al. 2011: no CDC42-directed GEF activity)
TIAM1 (Michiels et al. 1995: CDC42 directed GEF activity; Itoh et al. 2008, Jaiswal et al. 2013, Müller et al. 2020: no CDC42 directed GEF activity; Bagci et al. 2020: no binding to inactive CDC42)
TRIO (Peurois et al. 2017, Fortin et al. 2012, Jaiswal et al. 2013: CDC42 directed GEF activity, the presence of membrane may be necessary; Debant et al. 1996: no CDC42-directed GEF activity of either the N-terminal GEF1 domain or the C-terminal GEF2 domain of TRIO in vitro; Müller et al. 2020: no CDC42 directed GEF activity of the full-length TRIO; Bagci et al. 2020: no binding of full-length TRIO to inactive CDC42)
VAV2 (Itoh et al. 2008, Aoki et al. 2005, Heo et al. 2005, Jaiswal et al. 2013: CDC42-directed GEF activity; Bagci et al. 2020: binding to inactive CDC42; Müller et al. 2020: no CDC42 directed GEF activity)
VAV3 (Sachdev et al. 2002; Aoki et al. 2005: CDC42 directed GEF activity; Movilla and Bustelo 1999, Müller et al. 2020: no CDC42 directed GEF activity)
The following GEFs do not act on CDC42 or were shown to not bind to inactive CDC42 mutant in the high throughput screen by Bagci et al. 2020):
AKAP13 (Zheng et al. 1995; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ALS2 (Müller et al. 2020)
ARHGEF1 (Hart et al. 1996; Jaiswal et al. 2013; Jaiswal et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF2 (Krendel et al. 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF3 (Arthur et al. 2002; Müller et al. 2020)
ARHGEF7 (Manser et al. 1998; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF10L (Winkler et al. 2005; Müller et al. 2020)
ARHGEF17 (Rümenapp et al. 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ARHGEF18 (Niu et al. 2003; Blomquist et al. 2000; Müller et al. 2020)
ARHGEF28 (van Horck et al. 2001; Jaiswal et al. 2011; Jaiswal et al. 2013; Müller et al. 2020)
ARHGEF39 (Müller et al. 2020)
ARHGEF40 (Curtis et al. 2004; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK1 (Cote and Vuori 2002; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK2 (Kulkarni et al. 2011; Kwofie and Skowronski 2008; Müller et al. 2020)
DOCK3 (Kwofie and Skowronski 2008; Müller et al. 2020)
DOCK4 (Kwofie and Skowronski 2008; Abraham et al. 2015; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
DOCK5 (Vives et al. 2011; Müller et al. 2020; Bagci et al. 2020: no binding to inactive CDC42)
ECT2L (Müller et al. 2020)
FARP2 (Kubo et al. 2002; Müller et al. 2020)
FGD5 (Müller et al. 2020)
FGD6 (Müller et al. 2020)
ITSN2 (Müller et al. 2020)
KALRN (Penzes et al. 2001; Müller et al. 2020)
MCF2L2 (Müller et al. 2020)
NET1 (Alberts and Treisman 1998; Müller et al. 2020)
OBSCN (Ford Speelman et al. 2009)
PLEKHG5 (De Toledo et al. 2001; Müller et al. 2020)
PLEKHG6 (Müller et al. 2020)
PLEKHG7 (Müller et al. 2020)
RASGRF1 (Müller et al. 2020)
SOS1 (Nimnual et al. 1998; Müller et al. 2020)
SOS2 (Nimnual et al. 1998; Müller et al. 2020)
SWAP70 (Bagci et al. 2020: no binding to inactive CDC42)
TIAM2 (Müller et al. 2020)
VAV1 (Aghazadeh et al. 2000; Müller et al. 2020)
Reaction - small molecule participants:
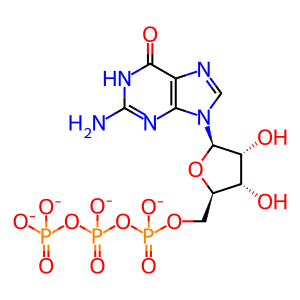
GTP [cytosol]
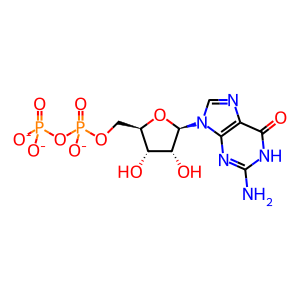
GDP [cytosol]
Reactome.org reaction link: R-HSA-9013159
======
Reaction input - small molecules:
GTP(4-)
Reaction output - small molecules:
GDP(3-)
Reactome.org link: R-HSA-9013159