Reaction: Dehydrogenase dehydrogenates 13-HDHA to 13-oxo-DHA
- in pathway: Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives
In activated macrophages, an unknown dehdyrogenase abstracts hydrogen from 13-hydroxy-docosahexaenoic acid to form the electrophilic oxo-derivative (EFOX) 13-oxo-DHA (Groeger et al. 2010). Potential candidates are cellular dehydrogenases such as 3α-hydroxysteroid dehydrogenases (3α-HSDs), which can convert 13- and 17-HDHA into their corresponding oxo-derivatives in the presence of NAD(P)+ in vitro (supplementary data, Groeger et al. 2010) or 5- and 15-hydroxyeicosanoid dehydrogenases (5- and 15-HEDH), which convert LOX products to 5-and 15-oxoETE (Erlemann et al. 2007). EFOXs can act as peroxisome proliferator-activated receptor-γ (PPARγ) agonists and inhibit pro-inflammatory cytokine and nitric oxide production, confirming their anti-inflammatory actions (Groeger et al. 2010).
Reaction - small molecule participants:
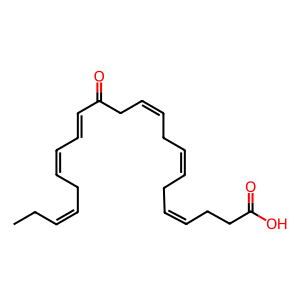
13-oxo-DHA [cytosol]
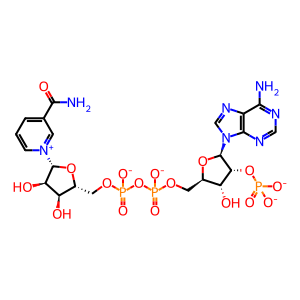
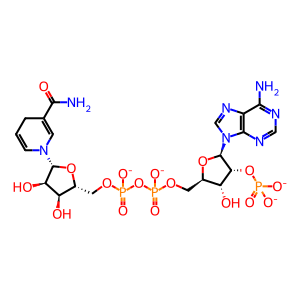
NADPH [cytosol]
NADP+ [cytosol]
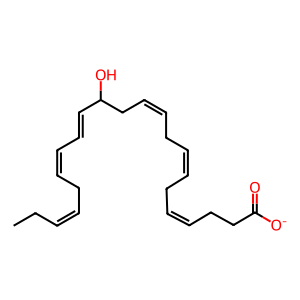
13-HDHA [cytosol]
Reactome.org reaction link: R-HSA-9027531
======
Reaction input - small molecules:
NADP(3-)
(4Z,7Z,10Z,14E,16Z,19Z)-13-hydroxydocosahexaenoate
Reaction output - small molecules:
(4Z,7Z,10Z,14E,16Z,19Z)-13-oxodocosahexaenoic acid
NADPH(4-)
Reactome.org link: R-HSA-9027531