Reaction: nsp16 acts as a cap 2'-O-methyltransferase to modify SARS-CoV-1 gRNA complement (minus strand)
- in pathway: Replication of the SARS-CoV-1 genome
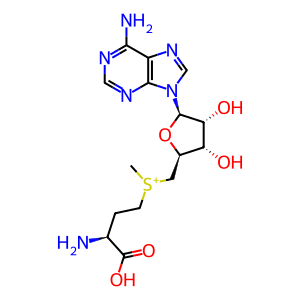
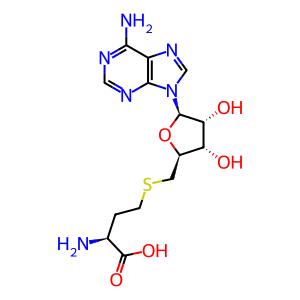
The genomic and subgenomic mRNAs of SARS-CoV-1 coronavirus, including the minus strand genomic RNA, are presumed to be capped at their 5′ end, based on studies of the mouse hepatitis virus (MHV) (Lai and Stohlman 1981) and the equine torovirus (van Vliet et al. 2002). Non-structural protein 16 (nsp16) acts as a 2'O-methyltransferase that converts coronavirus cap-0 to cap-1, which was first demonstrated with nsp16 cloned from the feline coronavirus (FCV) (Decroly et al. 2008). Cap-0 represents N7-methyl guanosine connected to the 5′ nucleotide through a 5′ to 5′ triphosphate linkage (also known as m7G cap or m7Gppp cap). Cap-1 is generated by an additional methylation on the 2′O position of the initiating nucleotide, and is also known as m7GpppNm. Non-structural protein 10 (nsp10) acts as an activator of nsp16 and is necessary for cap-1 synthesis (Bouvet et al. 2010, Decroly et al. 2011). Coronavirus RNAs with cap-1 are protected from IFIT-mediated interferon response. IFITs are interferon-induced proteins with tetratricopeptide repeats that recognize unmethylated 2'-O RNAs and act to inhibit expression of virally encoded mRNAs (Menachery et al. 2014).
Reaction - small molecule participants:
S-adenosyl-L-homocysteine [cytosol]
S-adenosyl-L-methionine [cytosol]
Reactome.org reaction link: R-HSA-9684030
======
Reaction input - small molecules:
S-adenosyl-L-methionine
Reaction output - small molecules:
S-adenosyl-L-homocysteine
Reactome.org link: R-HSA-9684030