Molecule: 1D-myo-inositol 1,4,5-trisphosphate(6-)
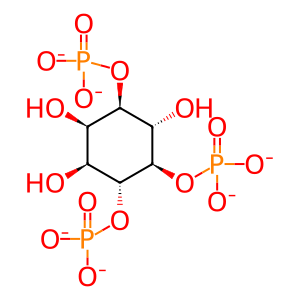
Hexaanion of 1D-myo-inositol 1,4,5-trisphosphate arising from glabal deprotonation of the phosphate OH groups; major species at pH 7.3.
Synonyms for 1D-myo-inositol 1,4,5-trisphosphate(6-) :
1D-myo-inositol 1,4,5-triphosphate hexaanion
1D-myo-inositol 1,4,5-triphosphate(6-)
1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate hexaanion
Molecular Formula: C6H9O15P3
Molecular wt: 414.04790 g/mole
Charge: -6
SMILES: O[C@@H]1[C@H](O)[C@@H](OP([O-])([O-])=O)[C@H](OP([O-])([O-])=O)[C@@H](O)[C@@H]1OP([O-])([O-])=O
InChIKey: MMWCIQZXVOZEGG-XJTPDSDZSA-H
InChI=1S/C6H15O15P3/c7-1-2(8)5(20-23(13,14)15)6(21-24(16,17)18)3(9)4(1)19-22(10,11)12/h1-9H,(H2,10,11,12)(H2,13,14,15)(H2,16,17,18)/p-6/t1-,2+,3+,4-,5-,6-/m1/s1
Sample reactions for this molecule:
